For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSGNAVRLQAINNVEAYVPPAVSFVAGEAPGEIFGSNVFTKAEMQARLPKAVFKSVLATIDKGARLDPAV ADAVAVAMKDWALEKGATHYAHVFYPMTGLTAEKHDSFLEPTADGQTMAEFAGKTLIQGEPDASSFPSGG LRSTFEARGYTGWDVTSPAYVLENPNGNTLCIPTIFVSMTGDALDYKTPLLRSQQAMGIHAERILKLFGH KELNKVVSFCGPEQEYFLVDRHFFVARPDLVNAGRTVFGAKPPKGQEFDDHYFGAVPERVLGFMMDTERE LFKLGIPAKTRHNEVAPGQFEVAPMFERANIASDHQQLLMTVFKTLAKKHGMECLFHEKPFAGVNGSGKH VNFSVGNAELGSLLVPGDTPHENAQFLVFCAAVIRAVHKYAGLLRVSVASATNDHRLGANEAPPAIISIF LGEQLADVFEQIAKGAATSSKGKGTMIIGVDTLPPLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVP MTILNTIMADSFDYMATVLEKAVEDGEDFDAAVQKLLTEIITEHGAVVFNGDGYSENWQIEAAERGLPNL KTTLDAIPELIKPESIDLFHKYDVFNERELHSRYEVRLEQYALTIAVEARLTLEMGTTVILPAAMRYQTE LAQNVAALKAAGVEPSMVALEAISAPLSDLSAALASLKMALSEHSADSALSEAKHAQEALLPAMDAVRCA ADTLESVVADDLWPLPTYQEMLYIL
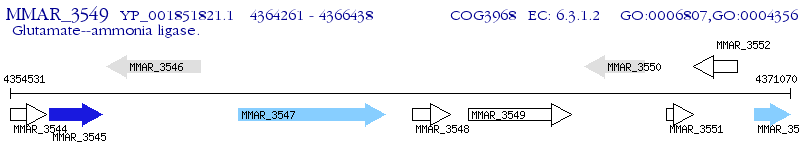
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3549 | - | - | 100% (725) | hypothetical protein MMAR_3549 |
| M. marinum M | MMAR_3294 | glnA2 | 1e-05 | 30.59% (85) | glutamine synthetase GlnA2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1910 | glnA3 | 6e-06 | 25.68% (183) | glutamine synthetase |
| M. gilvum PYR-GCK | Mflv_2596 | - | 0.0 | 86.34% (725) | glutamine synthetase, catalytic region |
| M. tuberculosis H37Rv | Rv1878 | glnA3 | 6e-06 | 25.68% (183) | glutamine synthetase |
| M. leprae Br4923 | MLBr_01631 | glnA2 | 2e-06 | 30.61% (98) | glutamine synthase class II |
| M. abscessus ATCC 19977 | MAB_1920 | - | 2e-05 | 29.73% (111) | glutamine synthetase |
| M. avium 104 | MAV_2829 | glnA | 5e-07 | 31.58% (133) | glutamine synthetase catalytic domain |
| M. smegmatis MC2 155 | MSMEG_6260 | glnT | 3e-06 | 23.36% (458) | glutamine synthetase, type III |
| M. thermoresistible (build 8) | TH_3396 | - | 3e-06 | 26.50% (117) | PUTATIVE glutamine synthetase, type I |
| M. ulcerans Agy99 | MUL_2782 | - | 0.0 | 98.76% (725) | hypothetical protein MUL_2782 |
| M. vanbaalenii PYR-1 | Mvan_4030 | - | 0.0 | 88.14% (725) | glutamine synthetase, catalytic region |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3549|M.marinum_M MSGNAVRLQAINNVEAYVPPAVSFVAGEAPGEIFGSNVFTKAEMQARLPK
MUL_2782|M.ulcerans_Agy99 MSGNAVRLQAINNVEAYVPPAVSFVAGEAPGEIFGSNVFTKAEMQARLPK
Mflv_2596|M.gilvum_PYR-GCK MSGNAVRLQAIDNVEAYVPPAITFDPAEAPGEIFGSNVFTKAEMQSRLPK
Mvan_4030|M.vanbaalenii_PYR-1 MSGNAVRLQAINNVEAYVPPAISFDPTEAPGQIFGSNVFTKAEMQLRLPK
Mb1910|M.bovis_AF2122/97 ----------------------------------MTATPLAAAAIAQLEA
Rv1878|M.tuberculosis_H37Rv ----------------------------------MTATPLAAAAIAQLEA
MAV_2829|M.avium_104 ----------------------------------MTGTPLAAAAIAQLEA
MAB_1920|M.abscessus_ATCC_1997 ------------------------------------MDRQKEFVLRTLEE
TH_3396|M.thermoresistible__bu ------------------------------------MDRQKEFVLRSLEE
MLBr_01631|M.leprae_Br4923 ------------------------------------MDRQKEFVLRTLEE
MSMEG_6260|M.smegmatis_MC2_155 ---------------------------------------MTSDLATLAEQ
MMAR_3549|M.marinum_M AVFKSVLATIDKGARLDPAVADAVAVAMKDWALEKGATHYAHVFYPMTGL
MUL_2782|M.ulcerans_Agy99 AVFKSVLATIDKGARLDPAVADAVAVAMKDWALEKGATHYAHVFYPMTGL
Mflv_2596|M.gilvum_PYR-GCK SVYKSVVATIEKGVKLDPAVADSVASAMKDWALSKGATHYAHVFYPMTGF
Mvan_4030|M.vanbaalenii_PYR-1 SVYKSVVATIEKGAKLDPAIADAVASAMKDWALSKGATHYAHVFYPMTGL
Mb1910|M.bovis_AF2122/97 EGVDTVIGTVVN--PAGLTQAKTVPIRRTNTFANPGLGASPVWHTFCIDQ
Rv1878|M.tuberculosis_H37Rv EGVDTVIGTVVN--PAGLTQAKTVPIRRTNTFANPGLGASPVWHTFCIDQ
MAV_2829|M.avium_104 EGVDTVIGTVVN--PAGLTLAKTVPIRRTNTFADPGLGASPSWHAFAVDQ
MAB_1920|M.abscessus_ATCC_1997 RDIRFVRLWFTD--VLGYLKSVSIAPAELEGAFEEGIGFD---GSSIEGF
TH_3396|M.thermoresistible__bu RDIRFVRLWFTD--VLGFLKSVAIAPAELEGAFEEGIGFD---GSAIEGF
MLBr_01631|M.leprae_Br4923 RDIRFVRLWFTD--VLGYLKSVAIAPAELEGAFEEGIGFD---GSSIEGF
MSMEG_6260|M.smegmatis_MC2_155 SGTKFILALFVD--LRGKPCAKLVPVEAIDMLATEGVGFA---GYAVGAM
: . . . : :. : *
MMAR_3549|M.marinum_M TAEKHDSFLEPTADGQTMAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
MUL_2782|M.ulcerans_Agy99 TAEKHDSFLEPTADGQTMAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
Mflv_2596|M.gilvum_PYR-GCK TAEKHDSFLEPVSDGQTLAEFAGKTLIQGEPDASSFPSGGLRTTFEARGY
Mvan_4030|M.vanbaalenii_PYR-1 TAEKHDSFLEPISDGETLAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
Mb1910|M.bovis_AF2122/97 CSIAFTADISVVGDQRLRIDLS----------------------------
Rv1878|M.tuberculosis_H37Rv CSIAFTADISVVGDQRLRIDLS----------------------------
MAV_2829|M.avium_104 TGIAFTAEVGVVGDQRLRIDLS----------------------------
MAB_1920|M.abscessus_ATCC_1997 ARVS-ESDTVAHPDPSTFQVLP----------------------------
TH_3396|M.thermoresistible__bu ARVS-EADMVARPDPSTFQVLP----------------------------
MLBr_01631|M.leprae_Br4923 ARVS-ESDTVAHPDPSTFQILP----------------------------
MSMEG_6260|M.smegmatis_MC2_155 GQEPKDPDLMALPDPASFTPIP----------------------------
. * :.
MMAR_3549|M.marinum_M TGWDVTSPAYVLENPNGNTLCIPTIFVSMTGDALDYKTPLLRSQQAMGIH
MUL_2782|M.ulcerans_Agy99 TGWDVTSPAYVLENPNGNTLCIPTIFVSMTGDALDYKTPLLRSQQAMGIH
Mflv_2596|M.gilvum_PYR-GCK TGWDVTSPAYILENPNGNTLCIPTVFVSMTGEALDFKTPLLRSQQAMGAQ
Mvan_4030|M.vanbaalenii_PYR-1 TGWDVTSPAYILENPNGNTLCIPTVFVSMTGEALDYKTPLLRSQQAMGVH
Mb1910|M.bovis_AF2122/97 ---------ALRIIGDG-LAWAPAGFFEQDGTPVPACSRGTLSRIEAALA
Rv1878|M.tuberculosis_H37Rv ---------ALRIIGDG-LAWAPAGFFEQDGTPVPACSRGTLSRIEAALA
MAV_2829|M.avium_104 ---------ALRIVGDG-LAWAPAAFFAQDGTPIPSCARGTLKRVEAALA
MAB_1920|M.abscessus_ATCC_1997 ---------WTTSAGKHHSARMFCDIKLPDGSPSWSDPRHVLRRQLAKAS
TH_3396|M.thermoresistible__bu ---------WTGDSGKHYSARMFCDITMPDGSPSWADPRHVLRRQLAKAG
MLBr_01631|M.leprae_Br4923 ---------WATTAGHHHSARMFCDITMPDGSSSWADPRHVLRRQLTKAN
MSMEG_6260|M.smegmatis_MC2_155 ---------FIKEG----LALVHCDPYVE-GKPWPYAPRVILKNLIQQAA
* . . .
MMAR_3549|M.marinum_M AERILKLFGHKELNKVVSFCGPEQEYFLVDRHFFVARPDLVNAGRTVFGA
MUL_2782|M.ulcerans_Agy99 AERILKLFGHEELNKVVSFCGPEQEYFLVDRHFFVARPELVNAGRTVFGA
Mflv_2596|M.gilvum_PYR-GCK AERILKLFGHSDFDNIVSFCGPEQEYFLVDRHFFLARPDLINAGRTLFGT
Mvan_4030|M.vanbaalenii_PYR-1 AERILALFGHDNLQKVVSFCGPEQEYFLVDRHFFLARPDLVNAGRTLFGA
Mb1910|M.bovis_AF2122/97 DA------------GIDAVIGHEVEFLLVDAD--------GQRLPSTLWA
Rv1878|M.tuberculosis_H37Rv DA------------GIDAVIGHEVEFLLVDAD--------GQRLPSTLWA
MAV_2829|M.avium_104 DA------------GIDAAIGHEVEFVLVAPD--------GQRLPSTLWA
MAB_1920|M.abscessus_ATCC_1997 DL------------GFSCYVHPEIEFFLLKNLPDDGTA--PIPADDAG--
TH_3396|M.thermoresistible__bu DQ------------GFTCYVHPEIEFFLLKPGPHDGSP--PVPADTSG--
MLBr_01631|M.leprae_Br4923 DL------------GFSCYVHPEIEFFLLKAGPEDGSMPIPVPVDNAG--
MSMEG_6260|M.smegmatis_MC2_155 DA------------GFEPWVGAEVEYFLLRRN-ADGSLVTADAADTAAQP
. * *:.*:
MMAR_3549|M.marinum_M KPPKGQEFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
MUL_2782|M.ulcerans_Agy99 KPPKGQGFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
Mflv_2596|M.gilvum_PYR-GCK RPPKGQEFDDHYFGAIPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
Mvan_4030|M.vanbaalenii_PYR-1 KPPKGQEFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPAQF
Mb1910|M.bovis_AF2122/97 ----------QYGVAGVLEHEAFVRDVNAAATAAGIAIEQFHPEYGANQF
Rv1878|M.tuberculosis_H37Rv ----------QYGVAGVLEHEAFVRDVNAAATAAGIAIEQFHPEYGANQF
MAV_2829|M.avium_104 ----------QYGLAGVLEHEAFVRDVIGSATRAGIAIEQLHPEYGANQF
MAB_1920|M.abscessus_ATCC_1997 ----------YFDQAVHEAAPNFRRHAIDALESMGISVEFSHHEGAPGQQ
TH_3396|M.thermoresistible__bu ----------YFDQSVHDTAPNFRRHAIEALEQMGISVEFSHHEGAPGQQ
MLBr_01631|M.leprae_Br4923 ----------YFDQAVHDSALNFRRHAIEALEFMGISVEFSHHEGAPGQQ
MSMEG_6260|M.smegmatis_MC2_155 ----------CYDARGLTRMYDHLTAISTAMNSLGWSNYANDHEDGNGQF
: . * . . * . *
MMAR_3549|M.marinum_M EVAPMFERANIASDHQQLLMTVFKTLAKKHGMECLFHEKPFAGVNGSGKH
MUL_2782|M.ulcerans_Agy99 EVAPMFERANIASDRQQLLMTVFKTLAKKHGMECLFHEKPFAGVNGSGKQ
Mflv_2596|M.gilvum_PYR-GCK EIAPMFERANIAADHQQLLMTTFKTIAKKHGMECLFHEKPFAGVNGSGKH
Mvan_4030|M.vanbaalenii_PYR-1 EVAPMFERANIASDHQQLLMTVFKTVAKKHGMECLFHEKPFAGVNGSGKH
Mb1910|M.bovis_AF2122/97 EISLAPQPPVAAADQLVLTRLIIGRTARRHGLRVSLSPAPFAGSIGSGAH
Rv1878|M.tuberculosis_H37Rv EISLAPQPPVAAADQLVLTRLIIGRTARRHGLRVSLSPAPFAGSIGSGAH
MAV_2829|M.avium_104 ELSLAPQPPVAAADQLILMRLIIGRAARRHGFRISLSPAPFAGDVGSGAH
MAB_1920|M.abscessus_ATCC_1997 EIDLRYADALSMADNVMTFRNVVKEVAIIDGVHATFMPKPFSDHPGSAMH
TH_3396|M.thermoresistible__bu EIDLRYADALSMADNVMTFRYVVKQVAISDGVRASFMPKPFAEHPGSAMH
MLBr_01631|M.leprae_Br4923 EIDLRFADALSMADNVMTFRYVIKEVAIENGARASFMPKPFAQHPGSAMH
MSMEG_6260|M.smegmatis_MC2_155 EQNFEFADALTTADRVITLRYLLSMIAAERGMVATFMPKPFADRTGSGLH
* . :*. . * * : **: **. :
MMAR_3549|M.marinum_M VNFSVGNAELGSLLVPGDTPHENAQFLVFCAAVIRAVHKYAGLLRVSVAS
MUL_2782|M.ulcerans_Agy99 VNFSVGNAELGSLLVPGDTPHENAQFLVFCAAVIRAVHKYAELLRVSVAS
Mflv_2596|M.gilvum_PYR-GCK VNFSMGNSQFGSLLVPGDTPHENAQFLVFCAAVIRAVHKFAGLLRVSVAS
Mvan_4030|M.vanbaalenii_PYR-1 VNFSVGNADLGSLLVPGDTPHENAQFLVFCAAIIRAVHKFGGLLRVSVAS
Mb1910|M.bovis_AF2122/97 QHFSLTMS--EGMLFSGGTGA--AGMTSAGEATVAGVLRGLPDAQGILCG
Rv1878|M.tuberculosis_H37Rv QHFSLTMS--EGMLFSGGTGA--AGMTSAGEAAVAGVLRGLPDAQGILCG
MAV_2829|M.avium_104 QHFSLSMA--EGPLFSGGAGT--AGMTPAGENAVAGVIRGLPEAQGVLCG
MAB_1920|M.abscessus_ATCC_1997 THMSLFEG--DTNAFHNPDDP--LQLSDVGKSFIAGILEHANEISAVTNQ
TH_3396|M.thermoresistible__bu THMSLFEG--DTNAFHSPDDP--LQLSDVAKSFIAGILEHANEISAVTNQ
MLBr_01631|M.leprae_Br4923 THMSLFEG--DVNAFHSDDDP--LQLSDVGKSFIAGILEHASEISAVTNQ
MSMEG_6260|M.smegmatis_MC2_155 FHLSLTSG--GNPVFPSETDERGLGLSDTAYAFLAGILDHACALQSVVAP
::*: . . . : : .:
MMAR_3549|M.marinum_M ATNDHRLGANEAPPAIISIFLGEQLADVFEQIAKGAATSSKGKGTMIIGV
MUL_2782|M.ulcerans_Agy99 ATNDHRLGANEAPPAIISIFLGEQLADVFEQIAKGAATSSKGKGTMIIGV
Mflv_2596|M.gilvum_PYR-GCK ATNDHRLGANEAPPAIISIFLGDQLADVFEQIAKGAATSSKGKGSMIIGC
Mvan_4030|M.vanbaalenii_PYR-1 ATNDHRLGANEAPPAIISIFLGEQLADVFDQIAKGAATSSKGKGVMHIGV
Mb1910|M.bovis_AF2122/97 SIVSGLRMRPGNWAGIYACWGTENREAAVRFVKGGAGSAY----------
Rv1878|M.tuberculosis_H37Rv SIVSGLRMRPGNWAGIYACWGTENREAAVRFVKGGAGSAY----------
MAV_2829|M.avium_104 SIVSGLRMRPGNWAGAYACWGTENREAAVRFIVDGPGSPR----------
MAB_1920|M.abscessus_ATCC_1997 WVNSYKRLVQGGEAPTAASWGAANRSALVRVPMYTPNKSS----------
TH_3396|M.thermoresistible__bu WVNSYKRLVHGGEAPTAASWGAANRSALVRVPMYTPHKAS----------
MLBr_01631|M.leprae_Br4923 WVNSYKRLVVGGEAPTTASWGAANRSALVRVPMYTPHKTS----------
MSMEG_6260|M.smegmatis_MC2_155 TVNSYKRTG-ATSTASGASWAPRLPSYGGNDRTHYIRVPD----------
. . : : .
MMAR_3549|M.marinum_M DTLPPLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMTILNTIMAD
MUL_2782|M.ulcerans_Agy99 DTLPPLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMTILNTIMAD
Mflv_2596|M.gilvum_PYR-GCK DTLPILPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTINVPMIVLNTIMAD
Mvan_4030|M.vanbaalenii_PYR-1 DTLPTLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMIILNTIMAD
Mb1910|M.bovis_AF2122/97 ----------------------GGNVEVKVVDPS---ANPYLASAAILGL
Rv1878|M.tuberculosis_H37Rv ----------------------GGNVEVKVVDPS---ANPYLASAAILGL
MAV_2829|M.avium_104 ----------------------GGNVEVKIVDPS---ANPYLASAAILGL
MAB_1920|M.abscessus_ATCC_1997 ----------------------SRRIEVRSPDSA---CNPYLTYAVLLAA
TH_3396|M.thermoresistible__bu ----------------------SRRVEVRSPDSA---CNPYLTFAVLLAA
MLBr_01631|M.leprae_Br4923 ----------------------SRRVEVRSPDSA---CNPYLAFAVLLAA
MSMEG_6260|M.smegmatis_MC2_155 ----------------------NQRIEMRGGDGS---ANPYLAVAAALGA
. ..*.: . . * . :.
MMAR_3549|M.marinum_M SFDYMATVLEKAVEDGEDFDAAVQKLLTEIITEHGAVVFNGDGYSENWQI
MUL_2782|M.ulcerans_Agy99 SFDYMATVLEKAVEDGEDFDAAVQKLLTEIITEHGAVVFNGDGYSENWQV
Mflv_2596|M.gilvum_PYR-GCK SLDYMATVLETAVGDGADFDTAVQKLLTEIITEHGAVVFNGDGYSENWHI
Mvan_4030|M.vanbaalenii_PYR-1 SFDYMATALEKAVAGGVDFDSAVQTLLTEIITEHGAVVFNGDGYSENWQI
Mb1910|M.bovis_AF2122/97 ALD----GMKTKAVLPSETTVDPTQLSDVDRDRAGILRLAADQADAIAVL
Rv1878|M.tuberculosis_H37Rv ALD----GMKTKAVLPSETTVDPTQLSDVDRDRAGILRLAADQADAIAVL
MAV_2829|M.avium_104 ALD----GIKSQAALPPEVTVDPAALSDADRDRDGVVLLTDSAAEAIAAL
MAB_1920|M.abscessus_ATCC_1997 GLR----GVEQGYTLGPEAEDDVWGLTRAERQAMGYKELPSTLENALIAM
TH_3396|M.thermoresistible__bu GLR----GVEKGYVLGPEAEDNVWTLTPEERRAMGFKELPSSLGQALAEM
MLBr_01631|M.leprae_Br4923 GLR----GVEKGYVLGPQAEDNVWDLTPEERLAMGYRELPTSLDSALGAM
MSMEG_6260|M.smegmatis_MC2_155 GLD----GIKRSIDPGPVGSG-------------GTTALPPTLLHAVDAL
.: :: * : :
MMAR_3549|M.marinum_M EAAERGLPNLKTTLDAIPELIKPESIDLFHKYDVFNERELHSRYEVRLEQ
MUL_2782|M.ulcerans_Agy99 EAAERGLPNLKTTLDAIPELIKPESIDLFHKYDVFNERELHSRYEVRLEQ
Mflv_2596|M.gilvum_PYR-GCK EAAERGLPNLKTTLDAITELIKPEAVEVFEKYGVFSERELHSRYEVRLEQ
Mvan_4030|M.vanbaalenii_PYR-1 EAAERGLPNLKTTLDAIPELIKPAAVEVFERYGVFSERELHSRYEVRLEQ
Mb1910|M.bovis_AF2122/97 DSSKLLR---CILGDPVVDAVVAVRQLEHERYGDLDPAQLADKFRMAWSV
Rv1878|M.tuberculosis_H37Rv DSSKLLR---CILGDPVVDAVVAVRQLEHERYGDLDPAQLADKFRMAWSV
MAV_2829|M.avium_104 DDSELMR---HILGDPVVDMVVAVRRLEQERYGDLDPAQLADRFRMAWSL
MAB_1920|M.abscessus_ATCC_1997 EGSELVA---EALGEHVFDFFLRNKRAEWARYRSNVTPFELRAYLGL---
TH_3396|M.thermoresistible__bu ERSELVA---EALGEHVFDFFLRNKRAEWENYRSHVTPYELKNYLSL---
MLBr_01631|M.leprae_Br4923 EVSEFVA---EALGEHVFDFFLRNKRAEWANYRSHVSLYELKTYLSL---
MSMEG_6260|M.smegmatis_MC2_155 EADPVVTGVLDAAGEGVATYFANLKREEFFTYHGTVTPWEVDTYLTAF--
: : : . * :
MMAR_3549|M.marinum_M YALTIAVEARLTLEMGTTVILPAAMRYQTELAQNVAALKAAGVEPSMVAL
MUL_2782|M.ulcerans_Agy99 YALTIAVEARLTLEMGTTVILPAAMRYQTELAQNVAALKAAGVEPSMVAL
Mflv_2596|M.gilvum_PYR-GCK YALTIAVEAKLSLELGTTMILPAAVRYQTELAQNVAALKAAGVAADTTLL
Mvan_4030|M.vanbaalenii_PYR-1 YALTIAVEAKLALELGTTVILPAAIRYQTELAQNVATLKAAGMDADTTLL
Mb1910|M.bovis_AF2122/97 --------------------------------------------------
Rv1878|M.tuberculosis_H37Rv --------------------------------------------------
MAV_2829|M.avium_104 --------------------------------------------------
MAB_1920|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3396|M.thermoresistible__bu --------------------------------------------------
MLBr_01631|M.leprae_Br4923 --------------------------------------------------
MSMEG_6260|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_3549|M.marinum_M EAISAPLSDLSAALASLKMALSEHSADSALSEAKHAQEALLPAMDAVRCA
MUL_2782|M.ulcerans_Agy99 EAISAPLSDLSAALASLKMALSEHSADSALSDAKHAQEALLPAMDAVRCA
Mflv_2596|M.gilvum_PYR-GCK QEVSAPIAELTAALATLKSALSGHSAESALDEATHAQKALLPAMEAVRAA
Mvan_4030|M.vanbaalenii_PYR-1 QAVSAPIAELTTALGALKAALSGHSGETALDEATHAQQALLPAMEAVRTA
Mb1910|M.bovis_AF2122/97 --------------------------------------------------
Rv1878|M.tuberculosis_H37Rv --------------------------------------------------
MAV_2829|M.avium_104 --------------------------------------------------
MAB_1920|M.abscessus_ATCC_1997 --------------------------------------------------
TH_3396|M.thermoresistible__bu --------------------------------------------------
MLBr_01631|M.leprae_Br4923 --------------------------------------------------
MSMEG_6260|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_3549|M.marinum_M ADTLESVVADDLWPLPTYQEMLYIL
MUL_2782|M.ulcerans_Agy99 ADTLESVVADDLWPLPTYQKMLYIL
Mflv_2596|M.gilvum_PYR-GCK ADELEAVVADDLWPLPTYQEMLYIL
Mvan_4030|M.vanbaalenii_PYR-1 ADTLETVVADDLWPLPTYQEMLYIL
Mb1910|M.bovis_AF2122/97 -------------------------
Rv1878|M.tuberculosis_H37Rv -------------------------
MAV_2829|M.avium_104 -------------------------
MAB_1920|M.abscessus_ATCC_1997 -------------------------
TH_3396|M.thermoresistible__bu -------------------------
MLBr_01631|M.leprae_Br4923 -------------------------
MSMEG_6260|M.smegmatis_MC2_155 -------------------------