For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSGNAVRLQAINNVEAYVPPAVSFVAGEAPGEIFGSNVFTKAEMQARLPKAVFKSVLATIDKGARLDPAV ADAVAVAMKDWALEKGATHYAHVFYPMTGLTAEKHDSFLEPTADGQTMAEFAGKTLIQGEPDASSFPSGG LRSTFEARGYTGWDVTSPAYVLENPNGNTLCIPTIFVSMTGDALDYKTPLLRSQQAMGIHAERILKLFGH EELNKVVSFCGPEQEYFLVDRHFFVARPELVNAGRTVFGAKPPKGQGFDDHYFGAVPERVLGFMMDTERE LFKLGIPAKTRHNEVAPGQFEVAPMFERANIASDRQQLLMTVFKTLAKKHGMECLFHEKPFAGVNGSGKQ VNFSVGNAELGSLLVPGDTPHENAQFLVFCAAVIRAVHKYAELLRVSVASATNDHRLGANEAPPAIISIF LGEQLADVFEQIAKGAATSSKGKGTMIIGVDTLPPLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVP MTILNTIMADSFDYMATVLEKAVEDGEDFDAAVQKLLTEIITEHGAVVFNGDGYSENWQVEAAERGLPNL KTTLDAIPELIKPESIDLFHKYDVFNERELHSRYEVRLEQYALTIAVEARLTLEMGTTVILPAAMRYQTE LAQNVAALKAAGVEPSMVALEAISAPLSDLSAALASLKMALSEHSADSALSDAKHAQEALLPAMDAVRCA ADTLESVVADDLWPLPTYQKMLYIL
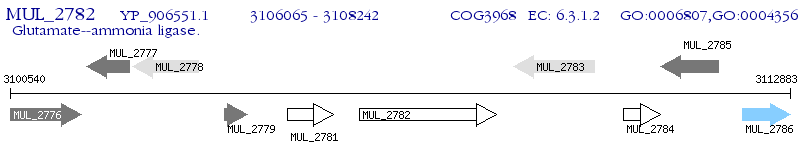
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2782 | - | - | 100% (725) | hypothetical protein MUL_2782 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2596 | - | 0.0 | 85.24% (725) | glutamine synthetase, catalytic region |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01631 | glnA2 | 2e-05 | 29.08% (141) | glutamine synthase class II |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3549 | - | 0.0 | 98.76% (725) | hypothetical protein MMAR_3549 |
| M. avium 104 | MAV_2829 | glnA | 3e-05 | 30.08% (133) | glutamine synthetase catalytic domain |
| M. smegmatis MC2 155 | MSMEG_6260 | glnT | 9e-06 | 23.36% (458) | glutamine synthetase, type III |
| M. thermoresistible (build 8) | TH_3396 | - | 8e-05 | 25.64% (117) | PUTATIVE glutamine synthetase, type I |
| M. vanbaalenii PYR-1 | Mvan_4030 | - | 0.0 | 87.03% (725) | glutamine synthetase, catalytic region |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2782|M.ulcerans_Agy99 MSGNAVRLQAINNVEAYVPPAVSFVAGEAPGEIFGSNVFTKAEMQARLPK
MMAR_3549|M.marinum_M MSGNAVRLQAINNVEAYVPPAVSFVAGEAPGEIFGSNVFTKAEMQARLPK
Mflv_2596|M.gilvum_PYR-GCK MSGNAVRLQAIDNVEAYVPPAITFDPAEAPGEIFGSNVFTKAEMQSRLPK
Mvan_4030|M.vanbaalenii_PYR-1 MSGNAVRLQAINNVEAYVPPAISFDPTEAPGQIFGSNVFTKAEMQLRLPK
MLBr_01631|M.leprae_Br4923 ---------------MDRQKEFVLRTLEERDIRFVRLWFTDVLGYLKSVA
TH_3396|M.thermoresistible__bu ---------------MDRQKEFVLRSLEERDIRFVRLWFTDVLGFLKSVA
MSMEG_6260|M.smegmatis_MC2_155 ------------------MTSDLATLAEQSGTKFILALFVDLRGKPCAKL
MAV_2829|M.avium_104 -------------MTGTPLAAAAIAQLEAEGVDTVIGTVVNPAGLTLAKT
* . ...
MUL_2782|M.ulcerans_Agy99 AVFKSVLATIDKGARLDPAVADAVAVAMKDWALEKGATHYAHVFYPMTGL
MMAR_3549|M.marinum_M AVFKSVLATIDKGARLDPAVADAVAVAMKDWALEKGATHYAHVFYPMTGL
Mflv_2596|M.gilvum_PYR-GCK SVYKSVVATIEKGVKLDPAVADSVASAMKDWALSKGATHYAHVFYPMTGF
Mvan_4030|M.vanbaalenii_PYR-1 SVYKSVVATIEKGAKLDPAIADAVASAMKDWALSKGATHYAHVFYPMTGL
MLBr_01631|M.leprae_Br4923 IAPAELEGAFEEGIGFDGSSIEGFARVS-ESDTVAHPDPSTFQILPWA--
TH_3396|M.thermoresistible__bu IAPAELEGAFEEGIGFDGSAIEGFARVS-EADMVARPDPSTFQVLPWT--
MSMEG_6260|M.smegmatis_MC2_155 VPVEAIDMLATEGVGFAGYAVGAMGQEPKDPDLMALPDPASFTPIPFI--
MAV_2829|M.avium_104 VPIRRTNTFADPGLGASPS-WHAFAVDQTGIAFTAEVGVVGDQRLRIDLS
* ...
MUL_2782|M.ulcerans_Agy99 TAEKHDSFLEPTADGQTMAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
MMAR_3549|M.marinum_M TAEKHDSFLEPTADGQTMAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
Mflv_2596|M.gilvum_PYR-GCK TAEKHDSFLEPVSDGQTLAEFAGKTLIQGEPDASSFPSGGLRTTFEARGY
Mvan_4030|M.vanbaalenii_PYR-1 TAEKHDSFLEPISDGETLAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
MLBr_01631|M.leprae_Br4923 ----------TTAGHHHSARMFCDITMPDGSSSWADPRHVLRRQLTKAND
TH_3396|M.thermoresistible__bu ----------GDSGKHYSARMFCDITMPDGSPSWADPRHVLRRQLAKAGD
MSMEG_6260|M.smegmatis_MC2_155 ----------KEG----LALVHCDPYVE-GKPWPYAPRVILKNLIQQAAD
MAV_2829|M.avium_104 ---------ALRIVGDGLAWAPAAFFAQDGTPIPSCARGTLKRVEAALAD
* . *:
MUL_2782|M.ulcerans_Agy99 TGWDVTSPAYVLENPNGNTLCIPTIFVSMTGDALDYKTPLLRSQQAMGIH
MMAR_3549|M.marinum_M TGWDVTSPAYVLENPNGNTLCIPTIFVSMTGDALDYKTPLLRSQQAMGIH
Mflv_2596|M.gilvum_PYR-GCK TGWDVTSPAYILENPNGNTLCIPTVFVSMTGEALDFKTPLLRSQQAMGAQ
Mvan_4030|M.vanbaalenii_PYR-1 TGWDVTSPAYILENPNGNTLCIPTVFVSMTGEALDYKTPLLRSQQAMGVH
MLBr_01631|M.leprae_Br4923 LGFSC---------------------------------------------
TH_3396|M.thermoresistible__bu QGFTC---------------------------------------------
MSMEG_6260|M.smegmatis_MC2_155 AGFEP---------------------------------------------
MAV_2829|M.avium_104 AGIDA---------------------------------------------
*
MUL_2782|M.ulcerans_Agy99 AERILKLFGHEELNKVVSFCGPEQEYFLVDRHFFVARPELVNAGRTVFGA
MMAR_3549|M.marinum_M AERILKLFGHKELNKVVSFCGPEQEYFLVDRHFFVARPDLVNAGRTVFGA
Mflv_2596|M.gilvum_PYR-GCK AERILKLFGHSDFDNIVSFCGPEQEYFLVDRHFFLARPDLINAGRTLFGT
Mvan_4030|M.vanbaalenii_PYR-1 AERILALFGHDNLQKVVSFCGPEQEYFLVDRHFFLARPDLVNAGRTLFGA
MLBr_01631|M.leprae_Br4923 ------------------YVHPEIEFFLLKAGPEDGSMPIPVPVDNAG--
TH_3396|M.thermoresistible__bu ------------------YVHPEIEFFLLKPGPHDGSP--PVPADTSG--
MSMEG_6260|M.smegmatis_MC2_155 ------------------WVGAEVEYFLLRRN-ADGSLVTADAADTAAQP
MAV_2829|M.avium_104 ------------------AIGHEVEFVLVAPDGQRLPSTLWAQYGLAG--
* *:.*:
MUL_2782|M.ulcerans_Agy99 KPPKGQGFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
MMAR_3549|M.marinum_M KPPKGQEFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
Mflv_2596|M.gilvum_PYR-GCK RPPKGQEFDDHYFGAIPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
Mvan_4030|M.vanbaalenii_PYR-1 KPPKGQEFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPAQF
MLBr_01631|M.leprae_Br4923 ----------YFDQAVHDSALNFRRHAIEALEFMGISVEFSHHEGAPGQQ
TH_3396|M.thermoresistible__bu ----------YFDQSVHDTAPNFRRHAIEALEQMGISVEFSHHEGAPGQQ
MSMEG_6260|M.smegmatis_MC2_155 ----------CYDARGLTRMYDHLTAISTAMNSLGWSNYANDHEDGNGQF
MAV_2829|M.avium_104 ----------------VLEHEAFVRDVIGSATRAGIAIEQLHPEYGANQF
. * . . * . *
MUL_2782|M.ulcerans_Agy99 EVAPMFERANIASDRQQLLMTVFKTLAKKHGMECLFHEKPFAGVNGSGKQ
MMAR_3549|M.marinum_M EVAPMFERANIASDHQQLLMTVFKTLAKKHGMECLFHEKPFAGVNGSGKH
Mflv_2596|M.gilvum_PYR-GCK EIAPMFERANIAADHQQLLMTTFKTIAKKHGMECLFHEKPFAGVNGSGKH
Mvan_4030|M.vanbaalenii_PYR-1 EVAPMFERANIASDHQQLLMTVFKTVAKKHGMECLFHEKPFAGVNGSGKH
MLBr_01631|M.leprae_Br4923 EIDLRFADALSMADNVMTFRYVIKEVAIENGARASFMPKPFAQHPGSAMH
TH_3396|M.thermoresistible__bu EIDLRYADALSMADNVMTFRYVVKQVAISDGVRASFMPKPFAEHPGSAMH
MSMEG_6260|M.smegmatis_MC2_155 EQNFEFADALTTADRVITLRYLLSMIAAERGMVATFMPKPFADRTGSGLH
MAV_2829|M.avium_104 ELSLAPQPPVAAADQLILMRLIIGRAARRHGFRISLSPAPFAGDVGSGAH
* . :*. : . * * : *** **. :
MUL_2782|M.ulcerans_Agy99 VNFSVGNAELGSLLVPGDTPHENAQFLVFCAAVIRAVHKYAELLRVSVAS
MMAR_3549|M.marinum_M VNFSVGNAELGSLLVPGDTPHENAQFLVFCAAVIRAVHKYAGLLRVSVAS
Mflv_2596|M.gilvum_PYR-GCK VNFSMGNSQFGSLLVPGDTPHENAQFLVFCAAVIRAVHKFAGLLRVSVAS
Mvan_4030|M.vanbaalenii_PYR-1 VNFSVGNADLGSLLVPGDTPHENAQFLVFCAAIIRAVHKFGGLLRVSVAS
MLBr_01631|M.leprae_Br4923 THMSLFEG--DVNAFHSDDDP--LQLSDVGKSFIAGILEHASEISAVTNQ
TH_3396|M.thermoresistible__bu THMSLFEG--DTNAFHSPDDP--LQLSDVAKSFIAGILEHANEISAVTNQ
MSMEG_6260|M.smegmatis_MC2_155 FHLSLTSG--GNPVFPSETDERGLGLSDTAYAFLAGILDHACALQSVVAP
MAV_2829|M.avium_104 QHFSLSMA--EGPLFSGGAGT--AGMTPAGENAVAGVIRGLPEAQGVLCG
::*: . . . : : .:
MUL_2782|M.ulcerans_Agy99 ATNDHRLGANEAPPAIISIFLGEQLADVFEQIAKGAATSSKGKGTMIIGV
MMAR_3549|M.marinum_M ATNDHRLGANEAPPAIISIFLGEQLADVFEQIAKGAATSSKGKGTMIIGV
Mflv_2596|M.gilvum_PYR-GCK ATNDHRLGANEAPPAIISIFLGDQLADVFEQIAKGAATSSKGKGSMIIGC
Mvan_4030|M.vanbaalenii_PYR-1 ATNDHRLGANEAPPAIISIFLGEQLADVFDQIAKGAATSSKGKGVMHIGV
MLBr_01631|M.leprae_Br4923 WVNSYKRLVVGGEAPTTASWG-----------------------------
TH_3396|M.thermoresistible__bu WVNSYKRLVHGGEAPTAASWG-----------------------------
MSMEG_6260|M.smegmatis_MC2_155 TVNSYKRTGAT-STASGASWA-----------------------------
MAV_2829|M.avium_104 SIVSGLRMRPGNWAGAYACWG-----------------------------
. . : :
MUL_2782|M.ulcerans_Agy99 DTLPPLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMTILNTIMAD
MMAR_3549|M.marinum_M DTLPPLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMTILNTIMAD
Mflv_2596|M.gilvum_PYR-GCK DTLPILPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTINVPMIVLNTIMAD
Mvan_4030|M.vanbaalenii_PYR-1 DTLPTLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMIILNTIMAD
MLBr_01631|M.leprae_Br4923 ---AANRSALVRVPMYTPHKTSSRRVEVRSPDSA---CNPYLAFAVLLAA
TH_3396|M.thermoresistible__bu ---AANRSALVRVPMYTPHKASSRRVEVRSPDSA---CNPYLTFAVLLAA
MSMEG_6260|M.smegmatis_MC2_155 ---PRLPSYGGNDRTHYIRVPDNQRIEMRGGDGS---ANPYLAVAAALGA
MAV_2829|M.avium_104 ---TENREAAVRFIVDGPGSPRGGNVEVKIVDPS---ANPYLASAAILGL
. . ..*.: . . * . :.
MUL_2782|M.ulcerans_Agy99 SFDYMATVLEKAVEDGEDFDAAVQKLLTEIITEHGAVVFNGDGYSENWQV
MMAR_3549|M.marinum_M SFDYMATVLEKAVEDGEDFDAAVQKLLTEIITEHGAVVFNGDGYSENWQI
Mflv_2596|M.gilvum_PYR-GCK SLDYMATVLETAVGDGADFDTAVQKLLTEIITEHGAVVFNGDGYSENWHI
Mvan_4030|M.vanbaalenii_PYR-1 SFDYMATALEKAVAGGVDFDSAVQTLLTEIITEHGAVVFNGDGYSENWQI
MLBr_01631|M.leprae_Br4923 GLR----GVEKGYVLGPQAEDNVWDLTPEERLAMGYRELPTSLDSALGAM
TH_3396|M.thermoresistible__bu GLR----GVEKGYVLGPEAEDNVWTLTPEERRAMGFKELPSSLGQALAEM
MSMEG_6260|M.smegmatis_MC2_155 GLD----GIKRSIDPGPVGSGGTTALPPTLLHAV----------DALEAD
MAV_2829|M.avium_104 ALD----GIKSQAALPPEVTVDPAALSDADRDRDGVVLLTDSAAEAIAAL
.: :: * .
MUL_2782|M.ulcerans_Agy99 EAAERGLPNLKTTLDAIPELIKPESIDLFHKYDVFNERELHSRYEVRLEQ
MMAR_3549|M.marinum_M EAAERGLPNLKTTLDAIPELIKPESIDLFHKYDVFNERELHSRYEVRLEQ
Mflv_2596|M.gilvum_PYR-GCK EAAERGLPNLKTTLDAITELIKPEAVEVFEKYGVFSERELHSRYEVRLEQ
Mvan_4030|M.vanbaalenii_PYR-1 EAAERGLPNLKTTLDAIPELIKPAAVEVFERYGVFSERELHSRYEVRLEQ
MLBr_01631|M.leprae_Br4923 EVSE-----------FVAEALGEHVFDFFLRN----KRAEWANYRSHVSL
TH_3396|M.thermoresistible__bu ERSE-----------LVAEALGEHVFDFFLRN----KRAEWENYRSHVTP
MSMEG_6260|M.smegmatis_MC2_155 PVVT-----------GVLDAAGEGVATYFANL----KREEFFTYHGTVTP
MAV_2829|M.avium_104 DDSE-----------LMRHILGDPVVDMVVAV-----RRLEQERYGDLDP
: . . * :
MUL_2782|M.ulcerans_Agy99 YALTIAVEARLTLEMGTTVILPAAMRYQTELAQNVAALKAAGVEPSMVAL
MMAR_3549|M.marinum_M YALTIAVEARLTLEMGTTVILPAAMRYQTELAQNVAALKAAGVEPSMVAL
Mflv_2596|M.gilvum_PYR-GCK YALTIAVEAKLSLELGTTMILPAAVRYQTELAQNVAALKAAGVAADTTLL
Mvan_4030|M.vanbaalenii_PYR-1 YALTIAVEAKLALELGTTVILPAAIRYQTELAQNVATLKAAGMDADTTLL
MLBr_01631|M.leprae_Br4923 YELKTYLSL-----------------------------------------
TH_3396|M.thermoresistible__bu YELKNYLSL-----------------------------------------
MSMEG_6260|M.smegmatis_MC2_155 WEVDTYLTAF----------------------------------------
MAV_2829|M.avium_104 AQLADRFRMAWSL-------------------------------------
: .
MUL_2782|M.ulcerans_Agy99 EAISAPLSDLSAALASLKMALSEHSADSALSDAKHAQEALLPAMDAVRCA
MMAR_3549|M.marinum_M EAISAPLSDLSAALASLKMALSEHSADSALSEAKHAQEALLPAMDAVRCA
Mflv_2596|M.gilvum_PYR-GCK QEVSAPIAELTAALATLKSALSGHSAESALDEATHAQKALLPAMEAVRAA
Mvan_4030|M.vanbaalenii_PYR-1 QAVSAPIAELTTALGALKAALSGHSGETALDEATHAQQALLPAMEAVRTA
MLBr_01631|M.leprae_Br4923 --------------------------------------------------
TH_3396|M.thermoresistible__bu --------------------------------------------------
MSMEG_6260|M.smegmatis_MC2_155 --------------------------------------------------
MAV_2829|M.avium_104 --------------------------------------------------
MUL_2782|M.ulcerans_Agy99 ADTLESVVADDLWPLPTYQKMLYIL
MMAR_3549|M.marinum_M ADTLESVVADDLWPLPTYQEMLYIL
Mflv_2596|M.gilvum_PYR-GCK ADELEAVVADDLWPLPTYQEMLYIL
Mvan_4030|M.vanbaalenii_PYR-1 ADTLETVVADDLWPLPTYQEMLYIL
MLBr_01631|M.leprae_Br4923 -------------------------
TH_3396|M.thermoresistible__bu -------------------------
MSMEG_6260|M.smegmatis_MC2_155 -------------------------
MAV_2829|M.avium_104 -------------------------