For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTARRIVSLAGPPMALLAVVIVAWYAVSYLVLDSSEKFLLPPPHDVVAQGLLDADTRDDITSALWVSAKV AIIGLVAAIVIGGVLAVAMAQAKWVERSLYPWVILLQTVPILAIVPVIGFWFGFELFARVLVVVIIALFP LIINPLQGLLGADRGLHDLFTLTGARPLTRLLKLQIPSAMPDVFVGLQSAAGLAVVGATVGDYFFGRGQI GLGMLLARYSSRLQSAEMLATVLTACLLGVVAFWIFGALGRRIVGRWSPAWGAR
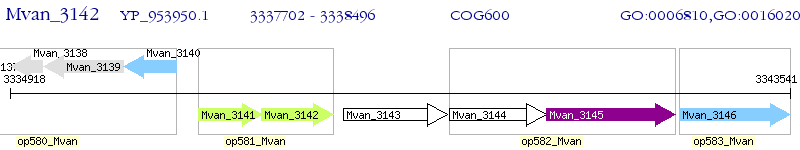
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3142 | - | - | 100% (264) | binding-protein-dependent transport systems inner membrane component |
| M. vanbaalenii PYR-1 | Mvan_3413 | - | 1e-17 | 27.08% (240) | binding-protein-dependent transport systems inner membrane |
| M. vanbaalenii PYR-1 | Mvan_0121 | - | 4e-16 | 23.55% (259) | binding-protein-dependent transport systems inner membrane |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3783c | proW | 2e-08 | 24.08% (191) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_3159 | - | 4e-15 | 27.73% (256) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv3757c | proW | 2e-08 | 24.08% (191) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2216 | - | 1e-16 | 27.47% (233) | sulfonate ABC transporter, permease protein SsuC |
| M. marinum M | MMAR_0035 | - | 9e-09 | 24.59% (183) | ABC transporter permease |
| M. avium 104 | MAV_2433 | - | 3e-17 | 24.79% (242) | putative aliphatic sulfonates transport permease protein SsuC |
| M. smegmatis MC2 155 | MSMEG_2635 | - | 1e-33 | 33.47% (239) | ABC transporter permease protein |
| M. thermoresistible (build 8) | TH_4654 | - | 6e-14 | 25.30% (249) | PUTATIVE putative binding-protein-dependent transport systems |
| M. ulcerans Agy99 | MUL_0034 | - | 1e-08 | 24.59% (183) | ABC transporter permease |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3159|M.gilvum_PYR-GCK MTAVAAPLTTP--RGPAAPAPELVDVIAGHRGR-RSIWG-RIPRPVRRMA
TH_4654|M.thermoresistible__bu VTSLQTARPAPRLRSPAQRRADVTDAQLLHRGEHRTRLGPGRAVPAALAV
MAB_2216|M.abscessus_ATCC_1997 MTTRNTLVAAAATDSDAP-----------ESGPSRREWVLERRWEIVRFL
MAV_2433|M.avium_104 MSIATALDAAPKRLGGAA-----------SGAPGG--LWARRKWEVVRLV
Mb3783c|M.bovis_AF2122/97 --------------------------------------------------
Rv3757c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0035|M.marinum_M --------------------------------------------------
MUL_0034|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_2635|M.smegmatis_MC2_155 MAVTASAGARPTGAQASSAAPGSAPRRRTRALFGTHPLHRTASALWRPLA
Mvan_3142|M.vanbaalenii_PYR-1 -----------------------------------MTARRIVSLAGPPMA
Mflv_3159|M.gilvum_PYR-GCK GPVAILAVWTIGSTSGLLNTD---LFPPPSDVAVTAWHLLTNGGLALHVG
TH_4654|M.thermoresistible__bu GPLLLVALWLITSQLGILDPR---TLPHPFDIARTAAELWSDGRLPANVA
MAB_2216|M.abscessus_ATCC_1997 SPLAVLLIWQAGSAWGLIDPD---ILPAPSTIARAGFELIANGQLAEALR
MAV_2433|M.avium_104 SPLALLALWQLGSAIGVIAQD---VLPAPSLILQAGVELTRNGQLADALH
Mb3783c|M.bovis_AF2122/97 ----------------------------------MHYLMTHPGAAWALTV
Rv3757c|M.tuberculosis_H37Rv ----------------------------------MHYLMTHPGAAWALTV
MMAR_0035|M.marinum_M ------------------------MEAATEFMGLYDFVVDRWSVLWFLAY
MUL_0034|M.ulcerans_Agy99 -------------------------------MGLYDFVVDRWSVLWFLAY
MSMEG_2635|M.smegmatis_MC2_155 LVVVLLAAWWAVTETELVAPY---ILPSPADTWLAAHDNAAY--LAHNTW
Mvan_3142|M.vanbaalenii_PYR-1 LLAVVIVAWYAVSYLVLDSSEKFLLPPPHDVVAQGLLDADTRDDITSALW
Mflv_3159|M.gilvum_PYR-GCK TSATRVLIGTALGITVGVVLAVLAGLTRTGEDLLDWSMQILKAVPNFALT
TH_4654|M.thermoresistible__bu ASLKLSLISLAIGVAAGLALALLAGLSRIGEALVDGPIQIKRAVPTLAII
MAB_2216|M.abscessus_ATCC_1997 VSGVRAAEGLLLGGVIGVVLGAAVGLSRLADAVVDPTMQMIRALPHLGLI
MAV_2433|M.avium_104 ISTVRVIEGLALGGVIGIVAGAAVGLSRWVEATVDPPLQMIRALPHLGLI
Mb3783c|M.bovis_AF2122/97 VHLRLSLLPVLIGLMSAVPLGLLVQRAPLLRRLTTATASVIFTIPSLALF
Rv3757c|M.tuberculosis_H37Rv VHLRLSLLPVLIGLMSAVPLGLLVQRAPLLRRLTTATASVIFTIPSLALF
MMAR_0035|M.marinum_M QHISLVGQTLILATAIALILGVLIYNSPLGVAAGNAVTAVGLTIPSYALL
MUL_0034|M.ulcerans_Agy99 QHISLVGQALILATAIALILGVLIYNSPLRVAAGNAVTAVGLTIPSYALL
MSMEG_2635|M.smegmatis_MC2_155 VTTWETVIGFVIAAIVGELVAVVMIYSAGVEKTVYPLILFAQVVPKIAIA
Mvan_3142|M.vanbaalenii_PYR-1 VSAKVAIIGLVAAIVIGGVLAVAMAQAKWVERSLYPWVILLQTVPILAIV
. . . : . .:* .:
Mflv_3159|M.gilvum_PYR-GCK PLLIIWMGIGEGPKIVLITMGVAIA---VYINTYSGIRGVDQQLVEMAQT
TH_4654|M.thermoresistible__bu PLAIIWFGIGDTMKIVIIATSVLIP---VYINTTAQLKGVDLRHVELAQT
MAB_2216|M.abscessus_ATCC_1997 PLFIVWFGIGELPKVLMVALGAAFP---LYLNTTSAIRQVDPKLFETAQV
MAV_2433|M.avium_104 PLFILWFGIGELPKVLLVALGVVFP---LYLNTFSAIRQVDPKMLETAQV
Mb3783c|M.bovis_AF2122/97 VVLPLIIGTRILDEANVIVALAAYTTALLVRAVLEALDAVPAQVHDAATA
Rv3757c|M.tuberculosis_H37Rv VVLPLIIGTRILDEANVIVALAAYTTALLVRAVLEALDAVPAQVHDAATA
MMAR_0035|M.marinum_M GVLVGIVGLGVLPSVIMLTFFGVFP---ILRNVVVGLTGVDKGLVESARG
MUL_0034|M.ulcerans_Agy99 GVLVGIVGLGVLPSVIMPTFFGVFP---ILRNVVVGLTGVDKGLVESARG
MSMEG_2635|M.smegmatis_MC2_155 PLFIVWLGFGPSPKILVAVLMAFFP---IVISGLAGLRSVDPEILELTST
Mvan_3142|M.vanbaalenii_PYR-1 PVIGFWFGFELFARVLVVVIIALFP---LIINPLQGLLGADRGLHDLFTL
: .* : . . : : . :
Mflv_3159|M.gilvum_PYR-GCK LEARRGTLITQVILPGAMPNFLVGLRLGLSSAWLSLIFAEMINTTEGIGF
TH_4654|M.thermoresistible__bu VGLSRVQFIRKVALPGALPGFFTGLRLAVTISWTALVVVEQVNATSGVGF
MAB_2216|M.abscessus_ATCC_1997 LGFSFWQRLRVIVIPGATPQVLVGLRQSLALSWLTLIVAEQINADAGVGF
MAV_2433|M.avium_104 LGFSFFQRFRRILVPGTAPQVLVGLRQSLAIAWLTLIVAEQINADKGIGF
Mb3783c|M.bovis_AF2122/97 IGYSRIAQMLKVELPLSIPVLVAGLRVVAVTNIAMVSVGSVIGIGGLGTW
Rv3757c|M.tuberculosis_H37Rv IGYSRIAQMLKVELPLSIPVLVAGLRVVAVTNIAMVSVGSVIGIGGLGTW
MMAR_0035|M.marinum_M MGMSRLTTLVRLELPLAWPVIMTGVRVSAQMLMGIAAIVAFALGPGLGGY
MUL_0034|M.ulcerans_Agy99 MGMSRLTTLVRLELPLAWPVIMTGVRVSAQMLMGIAAIVAFALGPGLGGY
MSMEG_2635|M.smegmatis_MC2_155 MGASRFKTFLKIRFPASLPQLMSGLKVAATLAVTGAVVGEFVGANEGLGY
Mvan_3142|M.vanbaalenii_PYR-1 TGARPLTRLLKLQIPSAMPDVFVGLQSAAGLAVVGATVGDYFFGRGQIGL
: : .* : * .. *:: .
Mflv_3159|M.gilvum_PYR-GCK LMSRAQ---TNLQFDVSLLVIVIYAVLGLASYALVRLLERLLLSWRNGFA
TH_4654|M.thermoresistible__bu LMTQAR---LYGQLEVVVVGLVVYAVFGLAGDHLVRTIERRALSWRRALG
MAB_2216|M.abscessus_ATCC_1997 LINNAR---DFLRIDVIIFGLVVYALLGIVTDAIVRVLERRALRYRS---
MAV_2433|M.avium_104 LINNAR---DFLRIDIIIFGLTIYALLGIITDAVVRLIERRAVRYRH---
Mb3783c|M.bovis_AF2122/97 FTAGYQ----TNKSDQIVAGVVAMFLLAIVVDVVINLAGRLATPWERAPR
Rv3757c|M.tuberculosis_H37Rv FTAGYQ----TNKSDQIVAGVVAMFLLAIVVDVVINLAGRLATPWERAPR
MMAR_0035|M.marinum_M IFSGISRMGGANATNSIVAATIGILILAVILDTVLNLIARLTTPRGIRA-
MUL_0034|M.ulcerans_Agy99 IFSGISRMGGANATNSIVAATIGILILAVILDTVLNVIARLTTPREIRA-
MSMEG_2635|M.smegmatis_MC2_155 VILQAN---GNVDTAMLFAALIIMSLLGIALFAVIEFAEKLLIPWHSSRR
Mvan_3142|M.vanbaalenii_PYR-1 GMLLAR-YSSRLQSAEMLATVLTACLLGVVAFWIFGALGRRIVGRWSPAW
. ::.: :. :
Mflv_3159|M.gilvum_PYR-GCK GFEATA-----------
TH_4654|M.thermoresistible__bu G----------------
MAB_2216|M.abscessus_ATCC_1997 -----------------
MAV_2433|M.avium_104 -----------------
Mb3783c|M.bovis_AF2122/97 AARRRRQVAAPITGGAR
Rv3757c|M.tuberculosis_H37Rv AARRRRQVAAPITGGAR
MMAR_0035|M.marinum_M -----------------
MUL_0034|M.ulcerans_Agy99 -----------------
MSMEG_2635|M.smegmatis_MC2_155 TVGSATSA---------
Mvan_3142|M.vanbaalenii_PYR-1 GAR--------------