For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HYLMTHPGAAWALTVVHLRLSLLPVLIGLMSAVPLGLLVQRAPLLRRLTTATASVIFTIPSLALFVVLPL IIGTRILDEANVIVALAAYTTALLVRAVLEALDAVPAQVHDAATAIGYSRIAQMLKVELPLSIPVLVAGL RVVAVTNIAMVSVGSVIGIGGLGTWFTAGYQTNKSDQIVAGVVAMFLLAIVVDVVINLAGRLATPWERAP RAARRRRQVAAPITGGAR*
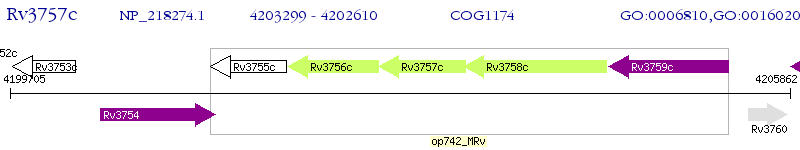
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3757c | proW | - | 100% (229) | POSSIBLE OSMOPROTECTANT (GLYCINE BETAINE/CARNITINE/CHOLINE/L-PROLINE) TRANSPORT INTEGRAL MEMBRANE PROTEIN ABC TRANSPORTER PROW |
| M. tuberculosis H37Rv | Rv3756c | proZ | 2e-16 | 26.22% (225) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. tuberculosis H37Rv | Rv0930 | pstA1 | 9e-09 | 32.39% (142) | phosphate ABC transporter transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3783c | proW | 1e-122 | 100.00% (229) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. gilvum PYR-GCK | Mflv_1209 | - | 3e-83 | 70.40% (223) | binding-protein-dependent transport systems inner membrane |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0263 | - | 5e-82 | 69.63% (214) | osmoprotectant ABC transporter ProW |
| M. marinum M | MMAR_5300 | proW | 5e-98 | 79.91% (229) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. avium 104 | MAV_0319 | - | 5e-91 | 74.24% (229) | ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_6332 | - | 1e-84 | 75.24% (210) | amino acid ABC transporter, permease protein |
| M. thermoresistible (build 8) | TH_0698 | - | 4e-22 | 31.98% (197) | ABC transporter, permease protein OpuCB |
| M. ulcerans Agy99 | MUL_4375 | proW | 2e-97 | 79.48% (229) | osmoprotectant (glycine betaine/carnitine/choline/L-proline) |
| M. vanbaalenii PYR-1 | Mvan_5599 | - | 5e-85 | 71.11% (225) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3757c|M.tuberculosis_H37Rv ---MHYLMTHPGAAWALTVVHLRLSLLPVLIGLMSAVPLGLLVQRAPLLR
Mb3783c|M.bovis_AF2122/97 ---MHYLMTHPGAAWALTVVHLRLSLLPVLIGLMSAVPLGLLVQRAPLLR
MMAR_5300|M.marinum_M ---MHYLMTHLGTAWALTMVHLRLSLIPVLIGTAIAIPLGLFVQHAPTLR
MUL_4375|M.ulcerans_Agy99 ---MHYLMTHLGTAWALTMVHLRLSLIPVLIGTAIAIPLGLFVQHAPTLR
MAV_0319|M.avium_104 ---MHYLLTHLDDAARLTVVHLRLSLVPVLIGLAIALPLGMLVQRRRFAR
Mflv_1209|M.gilvum_PYR-GCK ---MRYLFTHLDDLWVLTLIHLRLSLIPIVLGLLIAVPLGALVQRTTALR
Mvan_5599|M.vanbaalenii_PYR-1 ---MRYLVTHLDALWTLSLIHLRLSLIPILLGLLIAVPLGALVQRTTVLR
MSMEG_6332|M.smegmatis_MC2_155 ---MQYLLTHLDDLWELTVIHLRLSLVPIVLGLLIAVPLGALVQRTTTLR
MAB_0263|M.abscessus_ATCC_1997 ---MRYLFTHLGDAWQLSLIHLRLSLVPILIGLAIAIPCGALLHRRRTVR
TH_0698|M.thermoresistible__bu MDFIEFVRERWSVLSFLAYQHMSLVVQTLLLSTLLAVAVGVMLYRSTWGR
:.:: : . *: *: * : .:::. *:. * :: : *
Rv3757c|M.tuberculosis_H37Rv RLTTATASVIFTIPSLALFVVLPLIIGTRILDEANVIVALAAYTTALLVR
Mb3783c|M.bovis_AF2122/97 RLTTATASVIFTIPSLALFVVLPLIIGTRILDEANVIVALAAYTTALLVR
MMAR_5300|M.marinum_M RLTTATASVVFTIPSLALFVVLPLIIKTRILQEANVIIALSAYTTALLVR
MUL_4375|M.ulcerans_Agy99 RLTTATASVVFTIPSLALFVVLPLIIKMRILQEANVIIALSAYTTALLVR
MAV_0319|M.avium_104 RLTTAVASVVFTIPSLALFVVLPMIIGTRILDEANVMVALTAYTAALLVR
Mflv_1209|M.gilvum_PYR-GCK RLTTVTASIIFTIPSLALFVVLPLIIPTRILDEANVIVALTLYTVALLVR
Mvan_5599|M.vanbaalenii_PYR-1 RLTTVTASIVFTIPSLALFVVLPLIIPTRILDEANVIVALTLYTVALLVR
MSMEG_6332|M.smegmatis_MC2_155 RLTTVAASIIFTIPSLALFVALPLIIPTRILDEANVIVALTLYTTALLVR
MAB_0263|M.abscessus_ATCC_1997 RVTTVIASIVFTIPSLALFVALPLIIPTRILDEANVMVALTLYTTALLIR
TH_0698|M.thermoresistible__bu SLGDSVTSLGLTVPSYALLGVLVGIFGVGVLPS---VVLLTFFGFLPILR
: :*: :*:** **: .* *: :* . :: *: : ::*
Rv3757c|M.tuberculosis_H37Rv AVLEALDAVPAQVHDAATAIGYSRIAQMLKVELPLSIPVLVAGLRVVAVT
Mb3783c|M.bovis_AF2122/97 AVLEALDAVPAQVHDAATAIGYSRIAQMLKVELPLSIPVLVAGLRVVAVT
MMAR_5300|M.marinum_M AVLEALDAVPAQVRDAAVAVGYSPIARMLKVELPLSIPVLVSGLRVVVVT
MUL_4375|M.ulcerans_Agy99 AVLEALDAVPAQVRDAAVAVGYSPIARMLKVELPLSIPVLVSGLRVVVVT
MAV_0319|M.avium_104 AVLEALDAVPAQISDAATAVGYPRLTRMMKVELPLAVPVLIAGLRVVVVT
Mflv_1209|M.gilvum_PYR-GCK AVPEALDAVSPAVLDAATAVGYRPLTRMLKVELPLALPVLIASLRVVAVT
Mvan_5599|M.vanbaalenii_PYR-1 AVPEALDAVSPAVLDAATAVGYRPLTRMLKIELPLALPVLIASLRVVAVT
MSMEG_6332|M.smegmatis_MC2_155 AVPEALDAVPEHVRDAATAVGYRPLTRMLKVELPLSIPVLVASLRVVAVT
MAB_0263|M.abscessus_ATCC_1997 AVPEALDAIAPQVRDAATAVGYRPLARLVRVELPLSIPVLVAGLRVVAVT
TH_0698|M.thermoresistible__bu NVLVGLNGVDAGLVESARAMGMSRPATLLRLELPLAWPVIMTGVRISAQM
* .*:.: : ::* *:* : ::::****: **:::.:*: .
Rv3757c|M.tuberculosis_H37Rv NIAMVSVGSVIGIGGLGTWFTAGYQ----TNKSDQIVAGVVAMFLLAIVV
Mb3783c|M.bovis_AF2122/97 NIAMVSVGSVIGIGGLGTWFTAGYQ----TNKSDQIVAGVVAMFLLAIVV
MMAR_5300|M.marinum_M NIAMVSVGAVIGIGGLGTWFTAGYL----MDKSDQIVAGITVMFVLAILI
MUL_4375|M.ulcerans_Agy99 NIAMVSVGAVIGIGGLGTWFTAGYL----MDKSDQIVAGITVMFVLAILI
MAV_0319|M.avium_104 NIAMVSVGSVIGIGGLGSWFTQGYQ----TGKSDQILAGIIALFALAVAV
Mflv_1209|M.gilvum_PYR-GCK NISMVAVGSVIGIGGLGTWFTEGYQ----ANKSDQIIAGIVAIFVLAIVI
Mvan_5599|M.vanbaalenii_PYR-1 NISMVAVGSVIGIGGLGTWFTEGYQ----ANKSDQIIAGIVAIFVLAIVI
MSMEG_6332|M.smegmatis_MC2_155 NISMVSVGSVIGIGGLGTWFTEGYQ----SNKSSQIIAGIIAIFVLAIVV
MAB_0263|M.abscessus_ATCC_1997 NISMVSVGSVIGIGGLGTWFTEGYQ----ANKSSQIIAGIVAIFLLAVIV
TH_0698|M.thermoresistible__bu LMGIAAIAAFALGPGLGGYIFSGISRMGGANATNSIIVATVGILLLAVIL
:.:.::.:. *** :: * . :..*:.. :: **: :
Rv3757c|M.tuberculosis_H37Rv DVVINLAGRLATPWERAPRAARRRRQVAAPITGGAR
Mb3783c|M.bovis_AF2122/97 DVVINLAGRLATPWERAPRAARRRRQVAAPITGGAR
MMAR_5300|M.marinum_M DMVLNVAGRLATPWERSSRGRRRFR---SPIVGGAR
MUL_4375|M.ulcerans_Agy99 DMVLNVAGRLATPWERSSRGRRRFR---SPIVGGAR
MAV_0319|M.avium_104 DGLIVLGGRLVTPWAHAVRGGARRS-VVAPVVGGAR
Mflv_1209|M.gilvum_PYR-GCK DTLIMWLGKLLTPWTRAQPRSTRTQKAGASA-----
Mvan_5599|M.vanbaalenii_PYR-1 DTLIMLAGKALTPWTRAQPRATRAPRAEAGAPT---
MSMEG_6332|M.smegmatis_MC2_155 DTLIMLAGKLIAPWNRAEVTA---------------
MAB_0263|M.abscessus_ATCC_1997 DSLLVVLGRVATPWARATKTAGAT------------
TH_0698|M.thermoresistible__bu DAVLNLVTRLTTPRGIRV------------------
* :: : :*