For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGPASVRHLIDGEWLPGGGDALVSVNPARPTVVVAEGSCAVASDVDAAVGAAAAAAPSWAATGMHQRGAR LLAAAEIVERNAERWGLELATEEGKTRAEGVGEVRRAAQILRYYGNEGDRQAGEIYSSPRPGEQILVTRK PLGVVGVVTPFNFPIAIPAWKIAPALVYGNTVVWKPASTVPLLALRFAEALTDAGLPPGVLNLVVGGADI GDAIVNHPGIDGITFTGSTGVGRRIAAAAAARGVPAQAEMGGKNAAVVLDDADLDLAVEQVMLGAFRSTG QKCTATSRLIVTAGIADAFSEALLAQARALRVGDPVDDATQMGPVVSDSAQQSITAGIDTAVTQGATVLT GGRPYQDGALAEGYFVAPTVVELGAAPVDVWTDELFGPVLAIRRATDAEEAFALANDSEFGLSAAVFTQD MTRALQAVERIDVGVLHVNSESAGADPHVPFGGAKKSGLGPKEQGGAAREFFTHTTTVYLRGGRAGS*
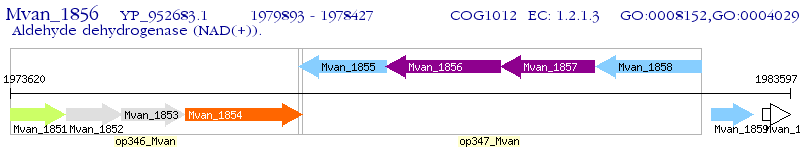
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1856 | - | - | 100% (488) | aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1328 | - | 1e-87 | 41.00% (478) | aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1572 | - | 3e-73 | 35.95% (459) | gamma-aminobutyraldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0775c | mmsA | 3e-76 | 35.99% (489) | methylmalonate-semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4860 | - | 1e-75 | 37.31% (453) | gamma-aminobutyraldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0753c | mmsA | 5e-77 | 35.99% (489) | methylmalonate-semialdehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 5e-59 | 31.94% (454) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0823 | - | 0.0 | 73.33% (480) | aldehyde dehydrogenase |
| M. marinum M | MMAR_1079 | mmsA | 6e-79 | 36.29% (485) | methylmalonate semialdehyde dehydrogenase, MmsA |
| M. avium 104 | MAV_2608 | - | 5e-85 | 40.63% (443) | aldehyde dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_6687 | - | 0.0 | 78.51% (484) | aldehyde dehydrogenase, thermostable |
| M. thermoresistible (build 8) | TH_2909 | - | 8e-78 | 35.46% (485) | PUTATIVE methylmalonate-semialdehyde dehydrogenase [acylating) |
| M. ulcerans Agy99 | MUL_0837 | mmsA | 9e-80 | 36.49% (485) | methylmalonate semialdehyde dehydrogenase, MmsA |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1856|M.vanbaalenii_PYR-1 -----MTGPASVRHLIDGEWLPGGGDALVSVNPARPTVVVAEGSCAVASD
MSMEG_6687|M.smegmatis_MC2_155 -----MT-PVTINHLVAGDWVAGAGDVVVSVNPARPGDIVAEGHNATTAQ
MAB_0823|M.abscessus_ATCC_1997 ------MHINEIQSFVGGRWISGGGQAVISVCPAFPDRVIAEGLAAGPAE
Mb0775c|M.bovis_AF2122/97 -------MTTQISHFIDGQRTAGQSTRSADVFDPNTGQIQAKVPMAGKSD
Rv0753c|M.tuberculosis_H37Rv -------MTTQISHFIDGQRTAGQSTRSADVFDPNTGQIQAKVPMAGKSD
MMAR_1079|M.marinum_M -------MTTQIPHFIDGRRTPGQSTRTADVLDPNTGQVQATVPMAAQAD
MUL_0837|M.ulcerans_Agy99 -------MTTQIPHFIDGRRTPGQSTRTADVLDPNTGQVQATVPMAAQAD
TH_2909|M.thermoresistible__bu -------MTRQIHHFINGQHTAGQSTRTADVMNPSTGKVQAKVVMGTAAD
Mflv_4860|M.gilvum_PYR-GCK ---------MTVASSWIDGAPVKTGGAQHTVINPATGESVAEFALAQPGD
MLBr_02573|M.leprae_Br4923 ---------MSIAT-----------------INPATGETVKTFTPTTQDE
MAV_2608|M.avium_104 MTDTATETLVQFDLLIGGESSSAASGLTYDSVDPFTGRPWARVPNAAAAD
. . . :
Mvan_1856|M.vanbaalenii_PYR-1 VDAAVGAAAAAAPS-WAATGMHQRGARLLAAAEIVERNAERWGLELATEE
MSMEG_6687|M.smegmatis_MC2_155 VDTAVAAAAEAFRA-WAATPTHQRGAVLLAAAQIVDRNAEAWGLELATEE
MAB_0823|M.abscessus_ATCC_1997 FDEAAAAARAAQPG-WAAMSLHARGAMLSHAAAVVDRNADAWGLELAAEE
Mb0775c|M.bovis_AF2122/97 IDAAVASAVEAQKG-WAAWNPQRRARVLMRFIELVNDTIDELAELLSREH
Rv0753c|M.tuberculosis_H37Rv IDAAVASAVEAQKG-WAAWNPQRRARVLMRFIELVNDTIDELAELLSREH
MMAR_1079|M.marinum_M IDAAVAGAAEAQIG-WAAWNPQRRARVLMRFIELVNQNTDELAELLSREH
MUL_0837|M.ulcerans_Agy99 IDAAVAGAAEAQVG-WAAWNPQRRARVLMRFIELVNQNTDELAELLSREH
TH_2909|M.thermoresistible__bu IDAAVAGAAEAQKE-WAAWNPQRRARVLMKFIDLVNQNVDELAELLSLEH
Mflv_4860|M.gilvum_PYR-GCK VDRAVVSARAALPG-WKGATPAERSAVLAKLAKLADDATADLVAEEVSQT
MLBr_02573|M.leprae_Br4923 VEDAIARAYARFDN-YRHTTFTQRAKWANATADLLEAEANQSAALMTLEM
MAV_2608|M.avium_104 VDRAVGAARAALNGPWGALTATARGKLLHRLGEIIAREADQLAELEVRDG
.: * * : *. : :
Mvan_1856|M.vanbaalenii_PYR-1 GKTRAEGVGEVRR-AAQILRYYGNEGDRQAGEIYSSPR--PGEQILVTRK
MSMEG_6687|M.smegmatis_MC2_155 GKTKAEGIGEVRR-AAQILRYYGNEGDRQAGEIFSSPR--AGEQILVTRK
MAB_0823|M.abscessus_ATCC_1997 GKTKAEGIAEVRR-AAQILRYYGNEGDRQTGEIFSSPR--RGEHILVTRK
Mb0775c|M.bovis_AF2122/97 GKTLADARGDVQR-GIEVIEFCLGIPHLLKGEYTEGAG--PGIDVYSLRQ
Rv0753c|M.tuberculosis_H37Rv GKTLADARGDVQR-GIEVIEFCLGIPHLLKGEYTEGAG--PGIDVYSLRQ
MMAR_1079|M.marinum_M GKTVPDARGDIQR-GIEVIEFCLGIPHLLKGEYTEGAG--PGIDVYSLRQ
MUL_0837|M.ulcerans_Agy99 GKTVPDARGDIQR-GIEVIEFCLGIPHLLKGEYTEGAG--PGIDVYSLRQ
TH_2909|M.thermoresistible__bu GKTLADAKGDIQR-GIEVIEFSIGIPHLQKGEYTEGAG--PGIDVYSMRQ
Mflv_4860|M.gilvum_PYR-GCK GKPVRLATEFDVPGSVDNIDFFAGAARHLEGKASAEYS--GDHTSSIRRE
MLBr_02573|M.leprae_Br4923 GKTLQSAKDEAMK-CAKGFRYYAEKAETLLADEPAEAAAVGASKALARYQ
MAV_2608|M.avium_104 GKLVREMVGQMRS-LPDYYFYYGGLADKLQGDVIPVDK--PNYFVYTRHE
** . : .. :
Mvan_1856|M.vanbaalenii_PYR-1 PLGVVGVVTPFNFPIAIPAWKIAPALVYGNTVVWKPASTVPLLALRFAEA
MSMEG_6687|M.smegmatis_MC2_155 PLGVVGVITPFNFPIAIPAWKIAPALVYGNTVVWKPASTVPVLAIRFAQA
MAB_0823|M.abscessus_ATCC_1997 PLGVVGVITPFNFPIAIPAWKIAPALVYGNAVLWKPASTVPLLAMRLAQA
Mb0775c|M.bovis_AF2122/97 PLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKPSERDPSVPVRLAEL
Rv0753c|M.tuberculosis_H37Rv PLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKPSERDPSVPVRLAEL
MMAR_1079|M.marinum_M PLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKPSERDPSVPLRLAEL
MUL_0837|M.ulcerans_Agy99 PLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKPSERDPSVPLRLAEL
TH_2909|M.thermoresistible__bu PLGVVAGITPFNFPAMIPLWKAGPALACGNAFILKPSERDPSVPVRLAEL
Mflv_4860|M.gilvum_PYR-GCK AVGVVATITPWNYPLQMAVWKVIPALAAGCSVVIKPAEITPLTTLTLARL
MLBr_02573|M.leprae_Br4923 PLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQCALYLADV
MAV_2608|M.avium_104 PVGVVGAITPWNSPLLLLTWKLAAGLAAGCTFVVKPSDHTPTSTLAFAKL
.:*** : *:* * ..* * : * :. * .: :*
Mvan_1856|M.vanbaalenii_PYR-1 LTDAGLPPGVLNLVVGG-ADIGDAIVNHPGIDGITFTGSTGVGRRIAAAA
MSMEG_6687|M.smegmatis_MC2_155 LTEAGLPAGALNLVIGD-SETGTAIVDHPRIDGLTFTGSTAVGRRLASVA
MAB_0823|M.abscessus_ATCC_1997 FDAAGLPPGVLNLVVGD-TGVGDAMVNDSRVDALTFTGSTGVGRRIAAAG
Mb0775c|M.bovis_AF2122/97 FIEAGLPAGVFQVVHGD-KEAVDAILHHPDIKAVGFVGSSDIAQYIYAGA
Rv0753c|M.tuberculosis_H37Rv FIEAGLPAGVFQVVHGD-KEAVDAILHHPDIKAVGFVGSSDIAQYIYAGA
MMAR_1079|M.marinum_M FVEAGLPPGVFQVVHGD-KEAVDAILNHPEIKAVGFVGSSDIAQYIYSGA
MUL_0837|M.ulcerans_Agy99 FVEAGLPPGVFQVVHGD-KEAVDAILNHPEIKAVGFVGSSDIAQYIYSGA
TH_2909|M.thermoresistible__bu FLQAGLPSGLFQVVHGD-KEAVDAILEHPTIQAVGFVGSSDIAQYIYAGA
Mflv_4860|M.gilvum_PYR-GCK ASEAGLPDGVLNVVTGSGADVGAALAGHADVDMVTFTGSTPVGRRVMLAA
MLBr_02573|M.leprae_Br4923 IARGGFPEDCFQTLLISANGVEAILR-DPRVAAATLTGSEPAGQAVGAIA
MAV_2608|M.avium_104 FAEAGFPPGVINVVTGWGPETGAALASHPGVDKVAFTGSTATGIHVGKAA
.*:* . :: : : .. : :.** . : .
Mvan_1856|M.vanbaalenii_PYR-1 AARGVPAQAEMGGKNAAVVLDDADLDLAVEQVMLGAFRSTGQKCTATSRL
MSMEG_6687|M.smegmatis_MC2_155 ATCGIPMQAEMGGKNAAVVLDDADLDVAVEQVMLGAFRSTGQKCTATSRL
MAB_0823|M.abscessus_ATCC_1997 AARGIPVQAEMGGKNAAVVLDDADVDLAVEQVLLGAFRSTGQKCTATSRL
Mb0775c|M.bovis_AF2122/97 AATGKRAQCFGGAKNHMIVMPDADLDQAVDALIGAGYGSAGERCMAISVA
Rv0753c|M.tuberculosis_H37Rv AATGKRAQCFGGAKNHMIVMPDADLDQAVDALIGAGYGSAGERCMAISVA
MMAR_1079|M.marinum_M AATGKRSQCFGGAKNHMIVMPDADLDQAVDALIGAGYGSAGERCMAISVA
MUL_0837|M.ulcerans_Agy99 AATGKRSQCFGGAKNHMIVMPDADLDQAVDALIGAGYGSTGERCMAISVA
TH_2909|M.thermoresistible__bu TAHGKRAQCFGGAKNHMIVMPDADLDQAVDALIGAGYGSAGERCMAISVA
Mflv_4860|M.gilvum_PYR-GCK AAHGHRTQLELGGKAPFVVFDDADLDAAIQGAVAGALINTGQDCTAATRA
MLBr_02573|M.leprae_Br4923 GNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRVQNNGQSCIAAKRF
MAV_2608|M.avium_104 IENMTRFSLELGGKSAQVVFADADLDAAANGVIAGVFAATGQTCLAGSRL
*.. :*: .**:* * : . *: * * .
Mvan_1856|M.vanbaalenii_PYR-1 IVT-AGIADAFSEALLAQARALRVGDPVDDATQMGPVVSDSAQQSITAGI
MSMEG_6687|M.smegmatis_MC2_155 IVT-DGIADKFTEALVERARALRVGDPTDDATEMGPVITDTSRASILAGI
MAB_0823|M.abscessus_ATCC_1997 VLT-EGIADAFLSLLKDRADALVVGDPTADDTQMGPVVSESAFGTITAGI
Mb0775c|M.bovis_AF2122/97 VPVGDQTAERLRARLIERINNLRVGHSLDPKADYGPLVTGAALARVRDYI
Rv0753c|M.tuberculosis_H37Rv VPVGDQTAERLRARLIERINNLRVGHSLDPKADYGPLVTGAALARVRDYI
MMAR_1079|M.marinum_M VPVGEQTADRLRAKLVERINNLRVGHSLDPKADYGPLVTEAALTRVRDYI
MUL_0837|M.ulcerans_Agy99 VPVGEQTADRLRAKLVERINNLRVGHSLDPKADYGPLVTEAALTRVRDYI
TH_2909|M.thermoresistible__bu VPVGDEIADRLRARLIERVANLRVGHSLDPKADYGPLVTEAALQRVRDYI
Mflv_4860|M.gilvum_PYR-GCK IVA-EGLYDDFVAGVAEVMSKVVIGDPADPDTDLGPLISAVHREKVAGMV
MLBr_02573|M.leprae_Br4923 IAH-ADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATESGRAQVEQQV
MAV_2608|M.avium_104 LVD-RSVADALVEKIVARAATIKLGDPKDPTTEMGPVSNLPQYEKVLSHF
: : : . : :*.. :: **: . : .
Mvan_1856|M.vanbaalenii_PYR-1 DTAVTQGATVLTGGRPYQ-----------DGALAEGYFVAPTVVELGAAP
MSMEG_6687|M.smegmatis_MC2_155 DTAVAQGATVLAGGRPYT-----------EGPLAEGYFIAPTVLELDGSP
MAB_0823|M.abscessus_ATCC_1997 GGAVAQGAVTLAGGHGYG-----------DGPLAAGYFIAPTIMELGMGA
Mb0775c|M.bovis_AF2122/97 GQGVAAGAELVVDGRDRASDDLTFGLPEGDANLEGGFFIGPTLFDHVAAH
Rv0753c|M.tuberculosis_H37Rv GQGVAAGAELVVDGRDRASDDLTFGLPEGDANLEGGFFIGPTLFDHVAAH
MMAR_1079|M.marinum_M GQGVASGAELVVDGRERASDDLAF----GDANLEGGYFIGPTLFDHVTTD
MUL_0837|M.ulcerans_Agy99 GQGVASGAELVVDGRERASDDLAF----GDANLEGGYFIGPTLFDHVTTD
TH_2909|M.thermoresistible__bu GQGVEAGAELLVDGRERGSDDLQF----GDDSLEGGYFIGPTLFDRVTTD
Mflv_4860|M.gilvum_PYR-GCK ERAPGEGGRVVTGGV-APDGP--------------GSFYRPTLIADVAES
MLBr_02573|M.leprae_Br4923 DAAAAAGAVIRCGGK-RPDRP--------------GWFYPPTVITDITKD
MAV_2608|M.avium_104 ASAREQGATVAYGGEPASDLG--------------GFFVKPTVLTGVDPS
. *. .* * * **:.
Mvan_1856|M.vanbaalenii_PYR-1 VDVWTDELFGPVLAIRRATDAEEAFALANDSEFGLSAAVFTQDMTRALQA
MSMEG_6687|M.smegmatis_MC2_155 AKVWSEELFGPVLAVRRAADADEAFELANDSEFGLSAAIFTQDVTRVLQA
MAB_0823|M.abscessus_ATCC_1997 VDLWTEELFGPVLAVRRAADTEEAFRLANDSEYGLSAAIFTKDLTRALSA
Mb0775c|M.bovis_AF2122/97 MSIYTDEIFGPVLCMVRARDYEEALRLPSEHEYGNGVAIFTRDGDAARDF
Rv0753c|M.tuberculosis_H37Rv MSIYTDEIFGPVLCMVRARDYEEALRLPSEHEYGNGVAIFTRDGDAARDF
MMAR_1079|M.marinum_M MSIYTDEIFGPVLCMVRAHDYEEALRLPSEHEYGNGVAVFTRDGDTARDF
MUL_0837|M.ulcerans_Agy99 MSIYTDEIFGPVLCMVRAHDYEEALRLPSEHEYGNGVAVFTRDGDTARDF
TH_2909|M.thermoresistible__bu MSIYRDEIFGPVLCMVRAKNYEEALALPSEHEYGNGVAIFTRDGDTARDF
Mflv_4860|M.gilvum_PYR-GCK SEVWRDEIFGPVLTVRSFTDDDDAIRQANDTKYGLAASAWTRDVYRAQRA
MLBr_02573|M.leprae_Br4923 MALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHF
MAV_2608|M.avium_104 MRAVAEEIFGPVLAVMTFEDEDEAVAAANGTEFGLAAAVWTKDVHRAHRV
:*:**** : : ::*. .. :* . :*::
Mvan_1856|M.vanbaalenii_PYR-1 VERIDVGVLHVNSESAGADPHVPFGGAKKSGLGPK-EQGGAAREFFTHTT
MSMEG_6687|M.smegmatis_MC2_155 MEQIDVGVLHVNSESAGADPHVPFGGDKKSGLGPK-EQGAAARDFFTHTT
MAB_0823|M.abscessus_ATCC_1997 VDHVDVGILHVNSESAGADPHVPFGGAKKSGLGPK-EQGRAAREFFTHTT
Mb0775c|M.bovis_AF2122/97 VSRGQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQHGPAAIQFYTKVK
Rv0753c|M.tuberculosis_H37Rv VSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQHGPAAIQFYTKVK
MMAR_1079|M.marinum_M VSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQHGPTSIQFYTKVK
MUL_0837|M.ulcerans_Agy99 VSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQHGPTSIQFYTKVK
TH_2909|M.thermoresistible__bu VSRVQCGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQHGPHSILFYTKTK
Mflv_4860|M.gilvum_PYR-GCK SREIHAGCVWIN-DHIPIISEMPHGGFGASGFGKD--MSQYSLEEYLSIK
MLBr_02573|M.leprae_Br4923 IDNIVAGQVFIN-GMTVSYPELPFGGIKRSGYGRE--LSTHGIREFCNIK
MAV_2608|M.avium_104 AAKLRAGTVWVN-AYRVVAPHVPFGGIGYSGIGRE--NGIDAVKDFTETK
. * : :* . ..** ** * . . : .
Mvan_1856|M.vanbaalenii_PYR-1 TVYLRGGRAGS-----------
MSMEG_6687|M.smegmatis_MC2_155 TVYLRGGRPGV-----------
MAB_0823|M.abscessus_ATCC_1997 TVYLRGEEAM------------
Mb0775c|M.bovis_AF2122/97 TVTSRWPSGIKDGAEFVIPTMS
Rv0753c|M.tuberculosis_H37Rv TVTSRWPSGIKDGAEFVIPTMS
MMAR_1079|M.marinum_M TVTSRWPSGIKDGAEFVIPTMK
MUL_0837|M.ulcerans_Agy99 TVTSRWPSGIKDGAEFVIPTMK
TH_2909|M.thermoresistible__bu TVTQRWPSGIKDGAEFSIPTMK
Mflv_4860|M.gilvum_PYR-GCK HVMSDITSAVDKDWHRTIFTKR
MLBr_02573|M.leprae_Br4923 TVWIA-----------------
MAV_2608|M.avium_104 AVWVELSGETRDPFTLG-----
*