For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHINEIQSFVGGRWISGGGQAVISVCPAFPDRVIAEGLAAGPAEFDEAAAAARAAQPGWAAMSLHARGAM LSHAAAVVDRNADAWGLELAAEEGKTKAEGIAEVRRAAQILRYYGNEGDRQTGEIFSSPRRGEHILVTRK PLGVVGVITPFNFPIAIPAWKIAPALVYGNAVLWKPASTVPLLAMRLAQAFDAAGLPPGVLNLVVGDTGV GDAMVNDSRVDALTFTGSTGVGRRIAAAGAARGIPVQAEMGGKNAAVVLDDADVDLAVEQVLLGAFRSTG QKCTATSRLVLTEGIADAFLSLLKDRADALVVGDPTADDTQMGPVVSESAFGTITAGIGGAVAQGAVTLA GGHGYGDGPLAAGYFIAPTIMELGMGAVDLWTEELFGPVLAVRRAADTEEAFRLANDSEYGLSAAIFTKD LTRALSAVDHVDVGILHVNSESAGADPHVPFGGAKKSGLGPKEQGRAAREFFTHTTTVYLRGEEAM
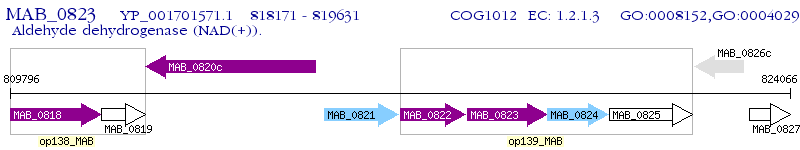
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0823 | - | - | 100% (486) | aldehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 1e-68 | 34.19% (468) | gamma-aminobutyraldehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1189c | - | 1e-67 | 34.03% (479) | methylmalonic acid semialdehyde dehydrogenase MmsA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0775c | mmsA | 1e-69 | 35.29% (493) | methylmalonate-semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4860 | - | 2e-71 | 34.97% (469) | gamma-aminobutyraldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0753c | mmsA | 2e-70 | 35.50% (493) | methylmalonate-semialdehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 1e-51 | 30.63% (431) | succinic semialdehyde dehydrogenase |
| M. marinum M | MMAR_1079 | mmsA | 6e-74 | 36.40% (489) | methylmalonate semialdehyde dehydrogenase, MmsA |
| M. avium 104 | MAV_1897 | - | 8e-74 | 36.43% (442) | betaine-aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6687 | - | 0.0 | 71.70% (477) | aldehyde dehydrogenase, thermostable |
| M. thermoresistible (build 8) | TH_3419 | - | 3e-71 | 36.63% (475) | PUTATIVE putative aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_0837 | mmsA | 8e-75 | 36.61% (489) | methylmalonate semialdehyde dehydrogenase, MmsA |
| M. vanbaalenii PYR-1 | Mvan_1856 | - | 0.0 | 73.33% (480) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1897|M.avium_104 MTDTTISNGVTEQLVHFDLVIGGESVAAESGRTYDSIDPYTALPWARVPD
TH_3419|M.thermoresistible__bu -----VQRTLPDKLPAPRLVIGGGDVISTSGGVYEHHYAATGGVQAEVPL
Mflv_4860|M.gilvum_PYR-GCK -------------MTVASSWIDGAPVKT-GGAQHTVINPATGESVAEFAL
MLBr_02573|M.leprae_Br4923 -------------MSIAT------------------INPATGETVKTFTP
MSMEG_6687|M.smegmatis_MC2_155 ----------MT-PVTINHLVAGDWVAGAGDVVVSVNPARPGDIVAEGHN
Mvan_1856|M.vanbaalenii_PYR-1 ----------MTGPASVRHLIDGEWLPGGGDALVSVNPARPTVVVAEGSC
MAB_0823|M.abscessus_ATCC_1997 -----------MHINEIQSFVGGRWISGGGQAVISVCPAFPDRVIAEGLA
Mb0775c|M.bovis_AF2122/97 ------------MTTQISHFIDGQRTAGQSTRSADVFDPNTGQIQAKVPM
Rv0753c|M.tuberculosis_H37Rv ------------MTTQISHFIDGQRTAGQSTRSADVFDPNTGQIQAKVPM
MMAR_1079|M.marinum_M ------------MTTQIPHFIDGRRTPGQSTRTADVLDPNTGQVQATVPM
MUL_0837|M.ulcerans_Agy99 ------------MTTQIPHFIDGRRTPGQSTRTADVLDPNTGQVQATVPM
. .
MAV_1897|M.avium_104 CGPADVDRAVAAARAALRGPWGQLTGTARGKLLWRLGELIAREAESLAEL
TH_3419|M.thermoresistible__bu AGPAEVDAAVAAARAAFP-AWRDLDLNRRRDILQELARRLVADGERLGAI
Mflv_4860|M.gilvum_PYR-GCK AQPGDVDRAVVSARAALP-GWKGATPAERSAVLAKLAKLADDATADLVAE
MLBr_02573|M.leprae_Br4923 TTQDEVEDAIARAYARFD-NYRHTTFTQRAKWANATADLLEAEANQSAAL
MSMEG_6687|M.smegmatis_MC2_155 ATTAQVDTAVAAAAEAFR-AWAATPTHQRGAVLLAAAQIVDRNAEAWGLE
Mvan_1856|M.vanbaalenii_PYR-1 AVASDVDAAVGAAAAAAP-SWAATGMHQRGARLLAAAEIVERNAERWGLE
MAB_0823|M.abscessus_ATCC_1997 AGPAEFDEAAAAARAAQP-GWAAMSLHARGAMLSHAAAVVDRNADAWGLE
Mb0775c|M.bovis_AF2122/97 AGKSDIDAAVASAVEAQK-GWAAWNPQRRARVLMRFIELVNDTIDELAEL
Rv0753c|M.tuberculosis_H37Rv AGKSDIDAAVASAVEAQK-GWAAWNPQRRARVLMRFIELVNDTIDELAEL
MMAR_1079|M.marinum_M AAQADIDAAVAGAAEAQI-GWAAWNPQRRARVLMRFIELVNQNTDELAEL
MUL_0837|M.ulcerans_Agy99 AAQADIDAAVAGAAEAQV-GWAAWNPQRRARVLMRFIELVNQNTDELAEL
:.: * * : *
MAV_1897|M.avium_104 EVRDGGKLVREMIG-QMKALPDYYIYYAGLADKLQGEVVPVDK--PNFFV
TH_3419|M.thermoresistible__bu ATLEVGTPNLLASS-SSAMAADWFTYYAGWIGRSAGEVVPVPA--GGALD
Mflv_4860|M.gilvum_PYR-GCK EVSQTGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKASAEYS--GDHTS
MLBr_02573|M.leprae_Br4923 MTLEMGKTLQSAKDEAMK-CAKGFRYYAEKAETLLADEPAEAAAVGASKA
MSMEG_6687|M.smegmatis_MC2_155 LATEEGKTKAEGIG-EVRRAAQILRYYGNEGDRQAGEIFSSPR--AGEQI
Mvan_1856|M.vanbaalenii_PYR-1 LATEEGKTRAEGVG-EVRRAAQILRYYGNEGDRQAGEIYSSPR--PGEQI
MAB_0823|M.abscessus_ATCC_1997 LAAEEGKTKAEGIA-EVRRAAQILRYYGNEGDRQTGEIFSSPR--RGEHI
Mb0775c|M.bovis_AF2122/97 LSREHGKTLADARG-DVQRGIEVIEFCLGIPHLLKGEYTEGAG--PGIDV
Rv0753c|M.tuberculosis_H37Rv LSREHGKTLADARG-DVQRGIEVIEFCLGIPHLLKGEYTEGAG--PGIDV
MMAR_1079|M.marinum_M LSREHGKTVPDARG-DIQRGIEVIEFCLGIPHLLKGEYTEGAG--PGIDV
MUL_0837|M.ulcerans_Agy99 LSREHGKTVPDARG-DIQRGIEVIEFCLGIPHLLKGEYTEGAG--PGIDV
: *. . : ..
MAV_1897|M.avium_104 YTRHEPVGVVGAITPWNSPLLLLTWKLAAGLAAGCTFVVKPSDHTPASTL
TH_3419|M.thermoresistible__bu YVLDEPLGVIAAIIPWNGPLISIGMKVAPALAAGNCVVLKPPENAPFSAL
Mflv_4860|M.gilvum_PYR-GCK SIRREAVGVVATITPWNYPLQMAVWKVIPALAAGCSVVIKPAEITPLTTL
MLBr_02573|M.leprae_Br4923 LARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQCAL
MSMEG_6687|M.smegmatis_MC2_155 LVTRKPLGVVGVITPFNFPIAIPAWKIAPALVYGNTVVWKPASTVPVLAI
Mvan_1856|M.vanbaalenii_PYR-1 LVTRKPLGVVGVVTPFNFPIAIPAWKIAPALVYGNTVVWKPASTVPLLAL
MAB_0823|M.abscessus_ATCC_1997 LVTRKPLGVVGVITPFNFPIAIPAWKIAPALVYGNAVLWKPASTVPLLAM
Mb0775c|M.bovis_AF2122/97 YSLRQPLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKPSERDPSVPV
Rv0753c|M.tuberculosis_H37Rv YSLRQPLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKPSERDPSVPV
MMAR_1079|M.marinum_M YSLRQPLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKPSERDPSVPL
MUL_0837|M.ulcerans_Agy99 YSLRQPLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKPSERDPSVPL
:.:**: : *:* * ..* * : * .. * .:
MAV_1897|M.avium_104 AFAKLFAEAGFPPGVINVVTGWGPQTGAALAAHPDVDKIAFTGSTATGIS
TH_3419|M.thermoresistible__bu RFAELAAEAGLPPGVLNVIPG-GAEAGSALCSHPGVDKITFTGGGETARK
Mflv_4860|M.gilvum_PYR-GCK TLARLASEAGLPDGVLNVVTGSGADVGAALAGHADVDMVTFTGSTPVGRR
MLBr_02573|M.leprae_Br4923 YLADVIARGGFPEDCFQTLLISANGVEAILR-DPRVAAATLTGSEPAGQA
MSMEG_6687|M.smegmatis_MC2_155 RFAQALTEAGLPAGALNLVIG-DSETGTAIVDHPRIDGLTFTGSTAVGRR
Mvan_1856|M.vanbaalenii_PYR-1 RFAEALTDAGLPPGVLNLVVG-GADIGDAIVNHPGIDGITFTGSTGVGRR
MAB_0823|M.abscessus_ATCC_1997 RLAQAFDAAGLPPGVLNLVVG-DTGVGDAMVNDSRVDALTFTGSTGVGRR
Mb0775c|M.bovis_AF2122/97 RLAELFIEAGLPAGVFQVVHG-DKEAVDAILHHPDIKAVGFVGSSDIAQY
Rv0753c|M.tuberculosis_H37Rv RLAELFIEAGLPAGVFQVVHG-DKEAVDAILHHPDIKAVGFVGSSDIAQY
MMAR_1079|M.marinum_M RLAELFVEAGLPPGVFQVVHG-DKEAVDAILNHPEIKAVGFVGSSDIAQY
MUL_0837|M.ulcerans_Agy99 RLAELFVEAGLPPGVFQVVHG-DKEAVDAILNHPEIKAVGFVGSSDIAQY
:* .*:* . :: : : .. : :.*. .
MAV_1897|M.avium_104 VGKAAIDNMTRFTLELGGKSAQVVFDDADLDAAAN-GVIAGVFAATGQTC
TH_3419|M.thermoresistible__bu VFAAAAQALTPVVLELGGKSANIVFPDADLDAAATMAVSAGLMMLSGQGC
Mflv_4860|M.gilvum_PYR-GCK VMLAAAAHGHRTQLELGGKAPFVVFDDADLDAAIQ-GAVAGALINTGQDC
MLBr_02573|M.leprae_Br4923 VGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVA-TAVTGRVQNNGQSC
MSMEG_6687|M.smegmatis_MC2_155 LASVAATCGIPMQAEMGGKNAAVVLDDADLDVAVE-QVMLGAFRSTGQKC
Mvan_1856|M.vanbaalenii_PYR-1 IAAAAAARGVPAQAEMGGKNAAVVLDDADLDLAVE-QVMLGAFRSTGQKC
MAB_0823|M.abscessus_ATCC_1997 IAAAGAARGIPVQAEMGGKNAAVVLDDADVDLAVE-QVLLGAFRSTGQKC
Mb0775c|M.bovis_AF2122/97 IYAGAAATGKRAQCFGGAKNHMIVMPDADLDQAVD-ALIGAGYGSAGERC
Rv0753c|M.tuberculosis_H37Rv IYAGAAATGKRAQCFGGAKNHMIVMPDADLDQAVD-ALIGAGYGSAGERC
MMAR_1079|M.marinum_M IYSGAAATGKRSQCFGGAKNHMIVMPDADLDQAVD-ALIGAGYGSAGERC
MUL_0837|M.ulcerans_Agy99 IYSGAAATGKRSQCFGGAKNHMIVMPDADLDQAVD-ALIGAGYGSTGERC
: . *.. :*: .**:* * . *: *
MAV_1897|M.avium_104 LAGSRLLVH-ESVADALVERIVARASTIKLGDPKDPATEMGPVSNQPQYE
TH_3419|M.thermoresistible__bu ILPTRLLVH-DDVYDEMVTRVVAGADALTVGDPWQPTTMMGPLATAAQLE
Mflv_4860|M.gilvum_PYR-GCK TAATRAIVA-EGLYDDFVAGVAEVMSKVVIGDPADPDTDLGPLISAVHRE
MLBr_02573|M.leprae_Br4923 IAAKRFIAH-ADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATESGRA
MSMEG_6687|M.smegmatis_MC2_155 TATSRLIVT-DGIADKFTEALVERARALRVGDPTDDATEMGPVITDTSRA
Mvan_1856|M.vanbaalenii_PYR-1 TATSRLIVT-AGIADAFSEALLAQARALRVGDPVDDATQMGPVVSDSAQQ
MAB_0823|M.abscessus_ATCC_1997 TATSRLVLT-EGIADAFLSLLKDRADALVVGDPTADDTQMGPVVSESAFG
Mb0775c|M.bovis_AF2122/97 MAISVAVPVGDQTAERLRARLIERINNLRVGHSLDPKADYGPLVTGAALA
Rv0753c|M.tuberculosis_H37Rv MAISVAVPVGDQTAERLRARLIERINNLRVGHSLDPKADYGPLVTGAALA
MMAR_1079|M.marinum_M MAISVAVPVGEQTADRLRAKLVERINNLRVGHSLDPKADYGPLVTEAALT
MUL_0837|M.ulcerans_Agy99 MAISVAVPVGEQTADRLRAKLVERINNLRVGHSLDPKADYGPLVTEAALT
. : : : . : :*.. : **: .
MAV_1897|M.avium_104 KVLSHFASAR--EQGATVAYGGEPAG------------ELGGFFVKPTVL
TH_3419|M.thermoresistible__bu RVTRAVDSALSDKAGKLLAGGRRPDG------------LDAGYFYRPTVF
Mflv_4860|M.gilvum_PYR-GCK KVAGMVERAPG-EGGRVVTGGVAPD--------------GPGSFYRPTLI
MLBr_02573|M.leprae_Br4923 QVEQQVDAAAA-AGAVIRCGGKRPD--------------RPGWFYPPTVI
MSMEG_6687|M.smegmatis_MC2_155 SILAGIDTAVAQGATVLAGGRPYT-----------EGPLAEGYFIAPTVL
Mvan_1856|M.vanbaalenii_PYR-1 SITAGIDTAVTQGATVLTGGRPYQ-----------DGALAEGYFVAPTVV
MAB_0823|M.abscessus_ATCC_1997 TITAGIGGAVAQGAVTLAGGHGYG-----------DGPLAAGYFIAPTIM
Mb0775c|M.bovis_AF2122/97 RVRDYIGQGVAAGAELVVDGRDRASDDLTFGLPEGDANLEGGFFIGPTLF
Rv0753c|M.tuberculosis_H37Rv RVRDYIGQGVAAGAELVVDGRDRASDDLTFGLPEGDANLEGGFFIGPTLF
MMAR_1079|M.marinum_M RVRDYIGQGVASGAELVVDGRERASDDLAF----GDANLEGGYFIGPTLF
MUL_0837|M.ulcerans_Agy99 RVRDYIGQGVASGAELVVDGRERASDDLAF----GDANLEGGYFIGPTLF
: . . * * **:.
MAV_1897|M.avium_104 TGVDRSMRAVAEEVFGPVLAVMTFTDEDEAIAAANDTEFGLAAGVWTKDV
TH_3419|M.thermoresistible__bu GDVDPASDLAQTEIFGPVLAILRFRSEEEAVELANNTQYGLAAYIHTRDL
Mflv_4860|M.gilvum_PYR-GCK ADVAESSEVWRDEIFGPVLTVRSFTDDDDAIRQANDTKYGLAASAWTRDV
MLBr_02573|M.leprae_Br4923 TDITKDMALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNE
MSMEG_6687|M.smegmatis_MC2_155 ELDGSPAKVWSEELFGPVLAVRRAADADEAFELANDSEFGLSAAIFTQDV
Mvan_1856|M.vanbaalenii_PYR-1 ELGAAPVDVWTDELFGPVLAIRRATDAEEAFALANDSEFGLSAAVFTQDM
MAB_0823|M.abscessus_ATCC_1997 ELGMGAVDLWTEELFGPVLAVRRAADTEEAFRLANDSEYGLSAAIFTKDL
Mb0775c|M.bovis_AF2122/97 DHVAAHMSIYTDEIFGPVLCMVRARDYEEALRLPSEHEYGNGVAIFTRDG
Rv0753c|M.tuberculosis_H37Rv DHVAAHMSIYTDEIFGPVLCMVRARDYEEALRLPSEHEYGNGVAIFTRDG
MMAR_1079|M.marinum_M DHVTTDMSIYTDEIFGPVLCMVRAHDYEEALRLPSEHEYGNGVAVFTRDG
MUL_0837|M.ulcerans_Agy99 DHVTTDMSIYTDEIFGPVLCMVRAHDYEEALRLPSEHEYGNGVAVFTRDG
*:**** : . ::*. .. :* . *::
MAV_1897|M.avium_104 HRAHRVAARLNAGTVWVN-AYRVVSPHVPFGGMGHSGIGRE--NGIDAVK
TH_3419|M.thermoresistible__bu RRAHVMAAALDAGGVAVN-GFPIVPSAAPFGGVKQSGFGRE--GGIWGLR
Mflv_4860|M.gilvum_PYR-GCK YRAQRASREIHAGCVWIN-DHIPIISEMPHGGFGASGFGKD--MSQYSLE
MLBr_02573|M.leprae_Br4923 SEQRHFIDNIVAGQVFIN-GMTVSYPELPFGGIKRSGYGRE--LSTHGIR
MSMEG_6687|M.smegmatis_MC2_155 TRVLQAMEQIDVGVLHVNSESAGADPHVPFGGDKKSGLGPK-EQGAAARD
Mvan_1856|M.vanbaalenii_PYR-1 TRALQAVERIDVGVLHVNSESAGADPHVPFGGAKKSGLGPK-EQGGAARE
MAB_0823|M.abscessus_ATCC_1997 TRALSAVDHVDVGILHVNSESAGADPHVPFGGAKKSGLGPK-EQGRAARE
Mb0775c|M.bovis_AF2122/97 DAARDFVSRGQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQHGPAAIQ
Rv0753c|M.tuberculosis_H37Rv DAARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQHGPAAIQ
MMAR_1079|M.marinum_M DTARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQHGPTSIQ
MUL_0837|M.ulcerans_Agy99 DTARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLNQHGPTSIQ
.* : :* . ..** ** * . .
MAV_1897|M.avium_104 DFTETKAVWVELSGATRDPFTLG-----
TH_3419|M.thermoresistible__bu EFQRTKNVYIGLQ---------------
Mflv_4860|M.gilvum_PYR-GCK EYLSIKHVMSDITSAVDKDWHRTIFTKR
MLBr_02573|M.leprae_Br4923 EFCNIKTVWIA-----------------
MSMEG_6687|M.smegmatis_MC2_155 FFTHTTTVYLR---GGRPGV--------
Mvan_1856|M.vanbaalenii_PYR-1 FFTHTTTVYLR---GGRAGS--------
MAB_0823|M.abscessus_ATCC_1997 FFTHTTTVYLR---GEEAM---------
Mb0775c|M.bovis_AF2122/97 FYTKVKTVTSRWPSGIKDGAEFVIPTMS
Rv0753c|M.tuberculosis_H37Rv FYTKVKTVTSRWPSGIKDGAEFVIPTMS
MMAR_1079|M.marinum_M FYTKVKTVTSRWPSGIKDGAEFVIPTMK
MUL_0837|M.ulcerans_Agy99 FYTKVKTVTSRWPSGIKDGAEFVIPTMK
: . *