For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDRASFDRLFDMTDRTVVVTGGTRGIGLALAEGYVLAGARVVVASRKADACEETARHLRELGGSAIGVPT HLGEVDDLGALVERTVAEFGGVDVLVNNAANPLAQPFGYMTVDALTKSFEVNLRGPVMLVQEALPHLKAS EHAAVLNMVSVGAFIFAPMLSIYASMKAAMMSFTRSMAAEFVHHGIRVNALAPGPVDTDMMRKNPQEAID GMVGGTLLGRLATPDEMVGAALLLTSDAGSYITGQVIIADGGGTPR
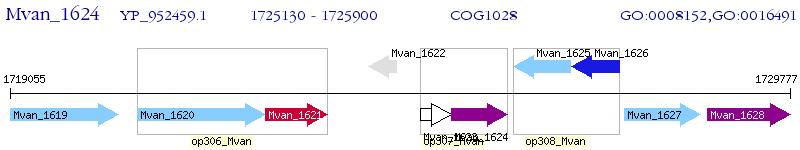
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1624 | - | - | 100% (256) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_5605 | - | 8e-37 | 38.65% (251) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_5156 | - | 2e-33 | 33.47% (248) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2788c | fabG | 3e-37 | 36.21% (243) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_4818 | - | 1e-133 | 92.97% (256) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2766c | fabG | 3e-37 | 36.21% (243) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 3e-23 | 30.67% (238) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0164 | - | 3e-88 | 63.24% (253) | short chain dehydrogenase/reductase |
| M. marinum M | MMAR_4927 | - | 9e-38 | 36.29% (248) | dehydrogenase/reductase |
| M. avium 104 | MAV_0714 | - | 7e-38 | 37.70% (244) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1740 | - | 1e-112 | 79.30% (256) | dehydrogenase/reductase SDR family protein member 4 |
| M. thermoresistible (build 8) | TH_2589 | - | 1e-112 | 79.69% (256) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4384 | - | 3e-37 | 36.95% (249) | short-chain type dehydrogenase/reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2788c|M.bovis_AF2122/97 -------------MTSLDLTGRTAIITGASRGIGLAIAQQLAAAGAHVVL
Rv2766c|M.tuberculosis_H37Rv -------------MTSLDLTGRTAIITGASRGIGLAIAQQLAAAGAHVVL
Mvan_1624|M.vanbaalenii_PYR-1 -------MDRASFDRLFDMTDRTVVVTGGTRGIGLALAEGYVLAGARVVV
Mflv_4818|M.gilvum_PYR-GCK -------MDRASFDRLFDMTDRTVVVTGGTRGIGLALAEGYALAGARVVV
MSMEG_1740|M.smegmatis_MC2_155 -------MDRQRFERLFDMSDRTVIVTGGTRGIGLSLAEGFVLAGARVVV
TH_2589|M.thermoresistible__bu -------VDRQTFDKLFDMTGRTVVVTGGTRGIGLALAEGYVLAGARVVV
MAB_0164|M.abscessus_ATCC_1997 -------MNLTDFQSLYDLSGRTAIVTGGTRGIGYAIAEALGACGASVVV
MUL_4384|M.ulcerans_Agy99 ---------MGYADQLFDLTDHVVLITGGSRGLGRQMAFAAARCGANVVI
MMAR_4927|M.marinum_M ---------------MGQFENKVAIVTGAAQGIGQAYADALAREGARVVV
MAV_0714|M.avium_104 ----------------MRFENKVGIVTGSGGGIGQAYAEALAREGAAVVV
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
: .: ::**. *:* * * *.:
Mb2788c|M.bovis_AF2122/97 TARRQEAADEAAAQVG----DRALGVGAHAVDEDAARRCVDLTLERFGSV
Rv2766c|M.tuberculosis_H37Rv TARRQEAADEAAAQVG----DRALGVGAHAVDEDAARRCVDLTLERFGSV
Mvan_1624|M.vanbaalenii_PYR-1 ASRKADACEETARHLR-ELGGSAIGVPTHLGEVDDLGALVERTVAEFGGV
Mflv_4818|M.gilvum_PYR-GCK ASRKADACEQAAHHLR-ELGAQAIGVPTHLGQVDDLGALVERTVAEFGGL
MSMEG_1740|M.smegmatis_MC2_155 ASRKPDACETAAAHLR-ELGGDAVGVPAHTGDVDDLGAVVERAVSEFGGV
TH_2589|M.thermoresistible__bu ASRKSDACERAAAHLR-ALGGDAIGVPTHLGDIDACEALVRRTVDEFGGI
MAB_0164|M.abscessus_ATCC_1997 SSRKSDACTAAAKELH-DKGYRALGVPAHMGELGDIDALVTATVDEYGGV
MUL_4384|M.ulcerans_Agy99 ASRNLDNCVATATEIESETGRSALAYQVHVGRWDQLDGLVAASYERFGKI
MMAR_4927|M.marinum_M ADINAEAAEKTAKQIV-ADGGTAIHVPVDVSDEESARAMADRTVSEFGGI
MAV_0714|M.avium_104 ADINAEAAEAVAKQIV-ADGGTAISVAVDVSDPESAKAMADRTLAEFGGI
MLBr_01807|M.leprae_Br4923 THR--------ASVVP----DWFFGVECDVTDNDAIGAAFKAVEEHQGPV
: * : . . . * :
Mb2788c|M.bovis_AF2122/97 DILINNAGTN--PAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMG
Rv2766c|M.tuberculosis_H37Rv DILINNAGTN--PAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMG
Mvan_1624|M.vanbaalenii_PYR-1 DVLVNNAAN---PLAQPFGYMTVDALTKSFEVNLRGPVMLVQEALPHLKA
Mflv_4818|M.gilvum_PYR-GCK DVLVNNAAN---PLAQPFGHMTVDALSKSFEVNLQGPVMLTQEALPHLKA
MSMEG_1740|M.smegmatis_MC2_155 DVVVNNAAN---ALAQPLGKMTPEAWAKSYEVNLRGPVFLVQHALPYLQK
TH_2589|M.thermoresistible__bu DVLVNNAAN---ALTQPLGELTVDAWTKSFEVNLRGPVFLVQAALPHLKT
MAB_0164|M.abscessus_ATCC_1997 DIVVNAAAN---PVAQPMGSYTPEALGKSFDVNVRGPVFLVQAALPHLTA
MUL_4384|M.ulcerans_Agy99 DTLINNAGMS--PLYDKLSDVTEKLFDAVLNLNLKGPFRLSALVGERMVA
MMAR_4927|M.marinum_M DYLVNNAAIYGGMKLDLLLTVPLDYYKKFMSVNHDGVLVCTRAVYKHMAK
MAV_0714|M.avium_104 DYLVNNAAIFGGMKLDFLLTIDPEYYKKFMSVNLDGALWCTRAVYKKMTK
MLBr_01807|M.leprae_Br4923 EVLVANAGIT---SDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIK
: :: *. : .:* . . .
Mb2788c|M.bovis_AF2122/97 EHGGAVVNTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-IRVN
Rv2766c|M.tuberculosis_H37Rv EHGGAVVNTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-IRVN
Mvan_1624|M.vanbaalenii_PYR-1 SEHAAVLNMVSVGAFIFAPMLSIYASMKAAMMSFTRSMAAEFVHHGIRVN
Mflv_4818|M.gilvum_PYR-GCK SDHAAVLNMVSVGAFIFAPMLSIYASMKAAMMSFTRSMAAEFVHDGIRVN
MSMEG_1740|M.smegmatis_MC2_155 SQNAAVLNMVSVGAFNFAPALSIYASSKAALMSVTRSMAAEFAPSGIRVN
TH_2589|M.thermoresistible__bu SPHAAVLNMVSVGAFLFSPGTAMYSAGKAALMSFTRSMAAQFAPLGIRVN
MAB_0164|M.abscessus_ATCC_1997 SEHASVLNVVSVAAFQYVPMLSMYAAMKATLMSFTRSMAAEYTARGIRVN
MUL_4384|M.ulcerans_Agy99 ADGGSIINVSSAGSLRPSADIIPYAAAKAGLNAMTEGLARAFGPT-VRVN
MMAR_4927|M.marinum_M RGGGAIVNQSSTAAWLYS---NFYGLAKVGVNGLTQQLSRELGGMKIRIN
MAV_0714|M.avium_104 RGGGAIVNQSSTAAWLYS---NYYGLAKVGINGLTQQLSRELGGRNIRIN
MLBr_01807|M.leprae_Br4923 QRFGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITAN
. : * . * * : .:. :: : *
Mb2788c|M.bovis_AF2122/97 AICPGVVRTRLAEA-LWKDHEDPLAATIALGRIGEPADIASAVAFLVSDA
Rv2766c|M.tuberculosis_H37Rv AICPGVVRTRLAEA-LWKDHEDPLAATIALGRIGEPADIASAVAFLVSDA
Mvan_1624|M.vanbaalenii_PYR-1 ALAPGPVDTDMMRK-NPQEAIDGMVGGTLLGRLATPDEMVGAALLLTSDA
Mflv_4818|M.gilvum_PYR-GCK ALAPGPVDTDMMRK-NPQEAIDGMVGGTLVGRLASADEMVGAALLLTSDA
MSMEG_1740|M.smegmatis_MC2_155 ALAPGPVDTDMMRK-NPQEAIDAMTAGTLMKRLAHPDEMVGAALLLCSDA
TH_2589|M.thermoresistible__bu ALAPGPVDTDMMRN-NPQEVVDMMAGATLMKRLATPDEMVGAALLLTSDA
MAB_0164|M.abscessus_ATCC_1997 ALAPGTVDTYMLQQ-NPQEIIDAMAAQSFMGRVAHTDEMIGPALLLLSDA
MUL_4384|M.ulcerans_Agy99 TLMAGPFLTDVSKAWNLDAATQNPFGHLALRRAGNPPEIVGAALFLASDA
MMAR_4927|M.marinum_M AIAPGPIDTEATRTVTPGEFVKDMVKQIPLSRMGTPEDLVGMCLFLLGDQ
MAV_0714|M.avium_104 AIAPGPIDTEANRTTTPKEMVDDIVKGLPLSRMGTPDDLVGMCLFLLSDE
MLBr_01807|M.leprae_Br4923 VVAPGFIDTEMTRA-MDSKYQEAALQAIPLGRIGAVEEIAGAVSFLASED
.: .* . * . . : * . :: . :* .:
Mb2788c|M.bovis_AF2122/97 ASWITGETMIIDGGLLLGNALGFRAAPSTEH
Rv2766c|M.tuberculosis_H37Rv ASWITGETMIIDGGLLLGNALGFRAAPSTEH
Mvan_1624|M.vanbaalenii_PYR-1 GSYITGQVIIADGGG-TPR------------
Mflv_4818|M.gilvum_PYR-GCK GSYITGQVIIADGGG-TPR------------
MSMEG_1740|M.smegmatis_MC2_155 GSYMTGQVVIVDGGG-TPR------------
TH_2589|M.thermoresistible__bu GSYITGTVLIADGGG-TPR------------
MAB_0164|M.abscessus_ATCC_1997 GSFITGQVIVADGGGGVQR------------
MUL_4384|M.ulcerans_Agy99 SSFTTGSILRADGGIP---------------
MMAR_4927|M.marinum_M ASWITGQIFNVDGGQIIRS------------
MAV_0714|M.avium_104 ASWITGQIFNVDGGQIIRS------------
MLBr_01807|M.leprae_Br4923 AGYISGAVIPVDGGLGMGH------------
..: :* . ***