For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGYADQLFDLTDHVVLITGGSRGLGRQMAFAAARCGANVVIASRNLDNCVATATEIESETGRSALAYQVH VGRWDQLDGLVAASYERFGKIDTLINNAGMSPLYDKLSDVTEKLFDAVLNLNLKGPFRLSALVGERMVAA DGGSIINVSSAGSLRPSADIIPYAAAKAGLNAMTEGLARAFGPTVRVNTLMAGPFLTDVSKAWNLDAATQ NPFGHLALRRAGNPPEIVGAALFLASDASSFTTGSILRADGGIP
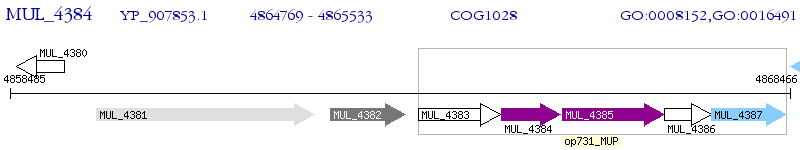
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4384 | - | - | 100% (254) | short-chain type dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_2173 | fabG | 1e-33 | 35.37% (246) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. ulcerans Agy99 | MUL_3898 | fabG | 6e-28 | 33.61% (238) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2788c | fabG | 6e-33 | 34.96% (246) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1202 | - | 1e-108 | 77.29% (251) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2766c | fabG | 6e-33 | 34.96% (246) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_02565 | fabG | 5e-18 | 29.55% (247) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_4913c | - | 4e-71 | 54.51% (244) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_5310 | - | 1e-139 | 98.03% (254) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_0311 | - | 1e-125 | 87.01% (254) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_6340 | - | 1e-114 | 79.13% (254) | short-chain dehydrogenase/reductase SDR |
| M. thermoresistible (build 8) | TH_3628 | - | 1e-112 | 79.37% (252) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_5605 | - | 1e-110 | 76.28% (253) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_4384|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5310|M.marinum_M --------------------------------------------------
MAV_0311|M.avium_104 --------------------------------------------------
MSMEG_6340|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1202|M.gilvum_PYR-GCK --------------------------------------------------
TH_3628|M.thermoresistible__bu --------------------------------------------------
Mvan_5605|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4913c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_4384|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5310|M.marinum_M --------------------------------------------------
MAV_0311|M.avium_104 --------------------------------------------------
MSMEG_6340|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1202|M.gilvum_PYR-GCK --------------------------------------------------
TH_3628|M.thermoresistible__bu --------------------------------------------------
Mvan_5605|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4913c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_4384|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5310|M.marinum_M --------------------------------------------------
MAV_0311|M.avium_104 --------------------------------------------------
MSMEG_6340|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1202|M.gilvum_PYR-GCK --------------------------------------------------
TH_3628|M.thermoresistible__bu --------------------------------------------------
Mvan_5605|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4913c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
Mb2788c|M.bovis_AF2122/97 --------------------------------------------------
Rv2766c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_4384|M.ulcerans_Agy99 --------------------------------------------------
MMAR_5310|M.marinum_M --------------------------------------------------
MAV_0311|M.avium_104 --------------------------------------------------
MSMEG_6340|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1202|M.gilvum_PYR-GCK --------------------------------------------------
TH_3628|M.thermoresistible__bu ------------------------------------------------VV
Mvan_5605|M.vanbaalenii_PYR-1 -----------------------------------------------MTA
MAB_4913c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Mb2788c|M.bovis_AF2122/97 -----MTSLDLTGRTAIITGASRGIGLAIAQQLAAAGAHVVLTARRQE--
Rv2766c|M.tuberculosis_H37Rv -----MTSLDLTGRTAIITGASRGIGLAIAQQLAAAGAHVVLTARRQE--
MUL_4384|M.ulcerans_Agy99 -MGYADQLFDLTDHVVLITGGSRGLGRQMAFAAARCGANVVIASRNLDNC
MMAR_5310|M.marinum_M -MGYADQLFDLTDHVVLITGGSRGLGRQMAFAAARCGADVVIASRNLDNC
MAV_0311|M.avium_104 -MGYAEQLFDLADRVVLITGGSRGLGREMAFGAARCGADVVIASRNLDNC
MSMEG_6340|M.smegmatis_MC2_155 -MGYADQLFDLTDRVVLITGGSRGLGREMAFGAARCGADVIIASRNLDSC
Mflv_1202|M.gilvum_PYR-GCK MTGFADRLFDLTDRVVLITGGSRGLGREMAVAAAHCGADVIIASRKYDAC
TH_3628|M.thermoresistible__bu HVGYADELFDLTDRVVLITGGSRGLGREMAMAAARCGADVIIASRKMDNC
Mvan_5605|M.vanbaalenii_PYR-1 SGGFADQLFDLTDRVVLVTGGSRGLGREMAFAAARCGADVVIASRKYEAC
MAB_4913c|M.abscessus_ATCC_199 ---------MLQDKVVVVTGGSRGLGRAMVRAFAAHGADVVIASRKIDSC
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAID--VESA
* .:..::**.:**:* :. * ** *: :
Mb2788c|M.bovis_AF2122/97 --AADEAAAQVGDRALGVGAHAVDEDAARRCVDLTLERFG-SVDILINNA
Rv2766c|M.tuberculosis_H37Rv --AADEAAAQVGDRALGVGAHAVDEDAARRCVDLTLERFG-SVDILINNA
MUL_4384|M.ulcerans_Agy99 VATATEIESETGRSALAYQVHVGRWDQLDGLVAASYERFG-KIDTLINNA
MMAR_5310|M.marinum_M VATATEIESETGRCAMAYQVHVGRWDQLDGLVAASYERFG-KIDTLINNA
MAV_0311|M.avium_104 VATAQQIEHETGRRAMAYQVHVGRWDQLDGLVEASYDRFG-KIDTLINNA
MSMEG_6340|M.smegmatis_MC2_155 VVTAEEIASETGRTAFPYSVHVGHWDELDGLVDAAYERFG-KVDVLVNNA
Mflv_1202|M.gilvum_PYR-GCK VATAEEITAQTGRAALPYGVHVGRWDELDGLVDAAYARFG-KVDVLVNNA
TH_3628|M.thermoresistible__bu VATAQEITEQTGRAALPYQVHVGRWDQLDGLVEAAYDRFG-KVDVLVNNA
Mvan_5605|M.vanbaalenii_PYR-1 VATADEIAAETGRAAFPYQVHVGRWDELDGLVDAAYDRFG-RVDVLVNNA
MAB_4913c|M.abscessus_ATCC_199 QELAAQVEAEHGRRALPVACNVSSWEQCDHLVDTVYREFG-RVDVLVNNA
MLBr_02565|M.leprae_Br4923 AEALDETANRVGGTTLSLDVTAD--DAVDKITEHLHDYQGGHADILVNNA
: . * :: . : . * * *:***
Mb2788c|M.bovis_AF2122/97 GTNPAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMGEHGGAVVNT
Rv2766c|M.tuberculosis_H37Rv GTNPAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMGEHGGAVVNT
MUL_4384|M.ulcerans_Agy99 GMSPLYDKLSDVTEKLFDAVLNLNLKGPFRLSALVGERMVAADGGSIINV
MMAR_5310|M.marinum_M GMSPLYDKLSDVTEKLFDAVLNLNLKGPFRLSALVGERMVAAGGGSIINV
MAV_0311|M.avium_104 GMSPLYDKLTDVTEKLFDAVVNLNLKGPFRLSALVGERMVAAGRGSIINV
MSMEG_6340|M.smegmatis_MC2_155 GMSPLYESLSAVTEKLYDAVFNLNLKGPFRLSALIGERMVADGGGRIINV
Mflv_1202|M.gilvum_PYR-GCK GMSPVYDKQSDVTEKLFDAVVNLNLKGPFRLSALIGERMVADGGGSIVNV
TH_3628|M.thermoresistible__bu GMSPVYDKQSDVTEQLFDAVVNLNLKGPFRLSALIGERMVADGGGSIINV
Mvan_5605|M.vanbaalenii_PYR-1 GMSPLYESLSAVTEKLFDSVVNLNLKGPFRLSVLVGDRMVADGGGSIINV
MAB_4913c|M.abscessus_ATCC_199 GLSPLYPSLDQVSEALFDKVIGVNLKGPFRLSASIATKMAAGAGGSIINI
MLBr_02565|M.leprae_Br4923 GIT-RDKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSISKGGRVIVL
* . . : :. :** .* : : * ::
Mb2788c|M.bovis_AF2122/97 ASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-IRVNAICPGVVRT
Rv2766c|M.tuberculosis_H37Rv ASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-IRVNAICPGVVRT
MUL_4384|M.ulcerans_Agy99 SSAGSLRPSADIIPYAAAKAGLNAMTEGLARAFGPT-VRVNTLMAGPFLT
MMAR_5310|M.marinum_M SSAGSLRPSADIIPYAAAKAGLNAMTEGLARAFGPT-VRVNTLMAGPFLT
MAV_0311|M.avium_104 STAGSLRPTPDIVPYAASKAGLNAMTEALAKAFGPA-VRVNTLMAGPFLT
MSMEG_6340|M.smegmatis_MC2_155 SSSGSLRPDQYMLPYAAAKAGLNVLTEGLAKAYGPT-VRVNTLMAGPFLT
Mflv_1202|M.gilvum_PYR-GCK STHGSLRPHPSFIPYAASKAGLNAMTEGLALAFGPS-VRVNTLMPGPFLT
TH_3628|M.thermoresistible__bu STHGSLRPHPTFIPYAASKAGLNVMTEGLALAFGPT-VRVNTLMPGPFLT
Mvan_5605|M.vanbaalenii_PYR-1 STHGSIRPHPSFVPYAASKAGLNAMTEALALAYGPT-VRVNTLMPGPFLT
MAB_4913c|M.abscessus_ATCC_199 SSIEAVRPDATALPYAAAKAGLNALTLGLSHTFGPT-VRTNTIQCGLFNT
MLBr_02565|M.leprae_Br4923 SSMAGIAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPGFIET
:: .: * ::**.: :* *: . : *:: * . *
Mb2788c|M.bovis_AF2122/97 RLAEALWKDHED-PLAATIALGRIGEPADIASAVAFLVSDAASWITGETM
Rv2766c|M.tuberculosis_H37Rv RLAEALWKDHED-PLAATIALGRIGEPADIASAVAFLVSDAASWITGETM
MUL_4384|M.ulcerans_Agy99 DVSKAWNLDAATQNPFGHLALRRAGNPPEIVGAALFLASDASSFTTGSIL
MMAR_5310|M.marinum_M DVSKAWNLDAATQNPFGHLTLRRAGNPPEIVGAALFLASDASSFTTGSIL
MAV_0311|M.avium_104 DVSRAWNLEAAQENPFRHLALQRAGDPREIVGAALFLASDASSFTTGSIL
MSMEG_6340|M.smegmatis_MC2_155 DVSRAWNMSGD---PFSHLALRRAGDPREIVGAALFLMSDASSFTTGSIV
Mflv_1202|M.gilvum_PYR-GCK DISNAWNFEGTD-NPFRGSALQRAGQPAEIVGAALFLMSDASSFTTGSIV
TH_3628|M.thermoresistible__bu DISKAWNLPEGS-NPFAGSALQRAGQPAEIVGAALFLMSDASSFTTGSIV
Mvan_5605|M.vanbaalenii_PYR-1 DISDAWTFTDGDTNPFGHSALQRAGQPAEIVGAALFLMSDASSFTTGSIV
MAB_4913c|M.abscessus_ATCC_199 DIAAAWP-EGFAEALIPSIPLRRVGEPEDIVGAALYLASEASAYCTGTTI
MLBr_02565|M.leprae_Br4923 QMTAAIPLATREVGRRLN-SLLQGGLPVDVAETIAYFAIPASNAVTGNVI
:: * .* : * * ::. : :: *: ** :
Mb2788c|M.bovis_AF2122/97 IIDGGLLLGNALGFRAAPSTEH
Rv2766c|M.tuberculosis_H37Rv IIDGGLLLGNALGFRAAPSTEH
MUL_4384|M.ulcerans_Agy99 RADGGIP---------------
MMAR_5310|M.marinum_M RADGGIP---------------
MAV_0311|M.avium_104 RADGGIP---------------
MSMEG_6340|M.smegmatis_MC2_155 RADGGLP---------------
Mflv_1202|M.gilvum_PYR-GCK RADGGSA---------------
TH_3628|M.thermoresistible__bu RADGGSV---------------
Mvan_5605|M.vanbaalenii_PYR-1 RADGGIP---------------
MAB_4913c|M.abscessus_ATCC_199 RLDGGIL---------------
MLBr_02565|M.leprae_Br4923 RVCGQAMLGA------------
*