For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRFENKVGIVTGSGGGIGQAYAEALAREGAAVVVADINAEAAEAVAKQIVADGGTAISVAVDVSDPESAK AMADRTLAEFGGIDYLVNNAAIFGGMKLDFLLTIDPEYYKKFMSVNLDGALWCTRAVYKKMTKRGGGAIV NQSSTAAWLYSNYYGLAKVGINGLTQQLSRELGGRNIRINAIAPGPIDTEANRTTTPKEMVDDIVKGLPL SRMGTPDDLVGMCLFLLSDEASWITGQIFNVDGGQIIRS
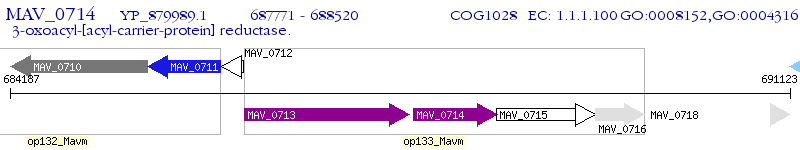
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0714 | - | - | 100% (249) | short chain dehydrogenase |
| M. avium 104 | MAV_4177 | - | 9e-41 | 38.96% (249) | putative short-chain type dehydrogenase/reductase y4lA |
| M. avium 104 | MAV_1395 | - | 1e-38 | 38.46% (247) | 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0792 | - | 1e-111 | 80.16% (247) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1601 | - | 1e-115 | 82.19% (247) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0769 | - | 1e-112 | 80.57% (247) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 2e-28 | 43.08% (195) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1219 | - | 1e-108 | 78.14% (247) | short chain dehydrogenase |
| M. marinum M | MMAR_4927 | - | 1e-115 | 83.47% (248) | dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_5858 | - | 1e-119 | 85.43% (247) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3037 | - | 1e-115 | 81.38% (247) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0478 | - | 1e-113 | 82.66% (248) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5156 | - | 1e-116 | 82.66% (248) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1601|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
MSMEG_5858|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0714|M.avium_104 --------------------------------------------------
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4927|M.marinum_M --------------------------------------------------
MUL_0478|M.ulcerans_Agy99 --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mflv_1601|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
MSMEG_5858|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0714|M.avium_104 --------------------------------------------------
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4927|M.marinum_M --------------------------------------------------
MUL_0478|M.ulcerans_Agy99 --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mflv_1601|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
MSMEG_5858|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0714|M.avium_104 --------------------------------------------------
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4927|M.marinum_M --------------------------------------------------
MUL_0478|M.ulcerans_Agy99 --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mflv_1601|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
MSMEG_5858|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0714|M.avium_104 --------------------------------------------------
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4927|M.marinum_M --------------------------------------------------
MUL_0478|M.ulcerans_Agy99 --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mflv_1601|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
MSMEG_5858|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0714|M.avium_104 --------------------------------------------------
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4927|M.marinum_M --------------------------------------------------
MUL_0478|M.ulcerans_Agy99 --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mflv_1601|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5156|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3037|M.thermoresistible__bu --------------------------------------------------
MSMEG_5858|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0714|M.avium_104 --------------------------------------------------
Mb0792|M.bovis_AF2122/97 --------------------------------------------------
Rv0769|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4927|M.marinum_M --------------------------------------------------
MUL_0478|M.ulcerans_Agy99 --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mflv_1601|M.gilvum_PYR-GCK ------------------MGLYGDQFNEKVAIVTGAGGGIGQAYAEALAR
Mvan_5156|M.vanbaalenii_PYR-1 ------------------MGLYGDQFKEKVAIVTGAGGGIGQAYAEALAR
TH_3037|M.thermoresistible__bu ------------------MG----QFDDKVAIVTGAGGGIGQAYAEALAR
MSMEG_5858|M.smegmatis_MC2_155 ------------------MGQYGTNFEDKVAIVTGAGGGIGQAYAEALAR
MAV_0714|M.avium_104 -----------------------MRFENKVGIVTGSGGGIGQAYAEALAR
Mb0792|M.bovis_AF2122/97 ------------------------MFDSKVAIVTGAAQGIGQAYAQALAR
Rv0769|M.tuberculosis_H37Rv ------------------------MFDSKVAIVTGAAQGIGQAYAQALAR
MMAR_4927|M.marinum_M ------------------MG----QFENKVAIVTGAAQGIGQAYADALAR
MUL_0478|M.ulcerans_Agy99 ------------------MG----QFENKVAIVTGAAQGIGQAYADALAR
MAB_1219|M.abscessus_ATCC_1997 ------------------MG----EFDNKVAIVTGAAQGIGEAYAHALAR
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
* ..: ***:. ***. * ****
Mflv_1601|M.gilvum_PYR-GCK EGAAVVVADINVEGAQKVADGIKGEGGNALAVRVDVSDPDSAKEMAAQAL
Mvan_5156|M.vanbaalenii_PYR-1 EGAAVVVADINTEGAQKVADGIKGEGGNALAVRVDVSDPESAKGMAAQTL
TH_3037|M.thermoresistible__bu EGAAVVVADINTEGAQKVADGIKGEGGDALAVPVDVSDPESAEEMAAQTV
MSMEG_5858|M.smegmatis_MC2_155 EGAAVVVADINLEGAQKVADAILGEGGNALAVRVDVSDIDSAKDMAAQTL
MAV_0714|M.avium_104 EGAAVVVADINAEAAEAVAKQIVADGGTAISVAVDVSDPESAKAMADRTL
Mb0792|M.bovis_AF2122/97 EGASVVVADINADGAAAVAKQIVADGGTVIHVPVDVSDEDSAKAMVDRAV
Rv0769|M.tuberculosis_H37Rv EGASVVVADINADGAAAVAKQIVADGGTAIHVPVDVSDEDSAKAMVDRAV
MMAR_4927|M.marinum_M EGARVVVADINAEAAEKTAKQIVADGGTAIHVPVDVSDEESARAMADRTV
MUL_0478|M.ulcerans_Agy99 EGARVVVADINAEAAEKTAKQIVADGGTAIHVPVDVSDEESARAMADRTV
MAB_1219|M.abscessus_ATCC_1997 EGAAVVVADINAEMGEGVAKQIVADGGRAIFAGVDVSDPDSAIAMADKAV
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
.** ***:**: . .* * ..** . :**** :. :. :.
Mflv_1601|M.gilvum_PYR-GCK SEFGGIDYLVNNAAIFGGMKLDFLITVDWDYYKKFMSVNLDGALVCTRAV
Mvan_5156|M.vanbaalenii_PYR-1 SEFGGIDYLVNNAAIFGGMKLDFLITVDWDYYKKFMSVNMDGALVCTRAV
TH_3037|M.thermoresistible__bu SEFGGIDYLVNNAAIFGGMKLDFLITVDWDYYKRFMSVNLDGALVCTRAV
MSMEG_5858|M.smegmatis_MC2_155 SEFGGIDYLVNNAAIFGGMKLDFLLTVDWDYYKKFMSVNLDGALVCTRAV
MAV_0714|M.avium_104 AEFGGIDYLVNNAAIFGGMKLDFLLTIDPEYYKKFMSVNLDGALWCTRAV
Mb0792|M.bovis_AF2122/97 GAFGGIDYLVNNAAIYGGMKLDLLLTVPLDYYKKFMSVNHDGVLVCTRAV
Rv0769|M.tuberculosis_H37Rv GAFGGIDYLVNNAAIYGGMKLDLLLTVPLDYYKKFMSVNHDGVLVCTRAV
MMAR_4927|M.marinum_M SEFGGIDYLVNNAAIYGGMKLDLLLTVPLDYYKKFMSVNHDGVLVCTRAV
MUL_0478|M.ulcerans_Agy99 SEFGGIDYLVNNAAIYGGMKLDLQLTVPLDYYKKFMSVNHDGVLVCTRAV
MAB_1219|M.abscessus_ATCC_1997 SEFGGIDYLVNNAAIYGGMKLDLLLSVPWDYYKKFMSVNQDGALVCTRAV
MLBr_00862|M.leprae_Br4923 AKHGVPDIVVNNAGIG---QAGRFLDTPPEEFDRVLAVNLGGVVNCCRAF
. .* * :****.* : . : : :.:.::** .*.: * **.
Mflv_1601|M.gilvum_PYR-GCK YRKMAKRG-GGAIVNQSSTAAWLYSN---FYGLAKVGINGLTQQLATELG
Mvan_5156|M.vanbaalenii_PYR-1 YRKMAKRG-GGAIVNQSSTAAWLYSN---FYGLAKVGINGLTQQLATELG
TH_3037|M.thermoresistible__bu YKKMAKRG-GGAIVNQSSTAAWLYSN---YYGLAKAGLNSLTQQLATELG
MSMEG_5858|M.smegmatis_MC2_155 YRKMAKRG-GGAIVNQSSTAAWLYSN---FYGLAKVGINGLTQQLSRELG
MAV_0714|M.avium_104 YKKMTKRG-GGAIVNQSSTAAWLYSN---YYGLAKVGINGLTQQLSRELG
Mb0792|M.bovis_AF2122/97 YKHMAKRG-GGAIVNQSSTAAWLYSN---FYGLAKVGVNGLTQQLARELG
Rv0769|M.tuberculosis_H37Rv YKHMAKRG-GGAIVNQSSTAAWLYSN---FYGLAKVGVNGLTQQLARELG
MMAR_4927|M.marinum_M YKHMAKRG-GGAIVNQSSTAAWLYSN---FYGLAKVGVNGLTQQLSRELG
MUL_0478|M.ulcerans_Agy99 YKHMAKRG-GGAIVNQSSTAAWLYSN---FYGLAKVGVNGLTQQLSRELG
MAB_1219|M.abscessus_ATCC_1997 YKHIAQRG-GGAIVNQSSTAAWLYSG---FYGLAKVGINGLTQQLSRELG
MLBr_00862|M.leprae_Br4923 GQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAELD
:::.:** ** *** ** **: * :*.. ::: * **.
Mflv_1601|M.gilvum_PYR-GCK GQNIRVNAIAPGPIDT---EANRTTTPQEMVADIVKG-------IPLSRM
Mvan_5156|M.vanbaalenii_PYR-1 GQNIRVNAIAPGPIDT---EANRTTTPQEMVADIVKG-------IPLSRM
TH_3037|M.thermoresistible__bu GQNIRVNAIAPGPIDT---EANRSTTPKEMVDDIVKR-------IPLSRL
MSMEG_5858|M.smegmatis_MC2_155 GQNIRINAIAPGPIDT---EANRTTTPKEMVDDIVKG-------IPLSRM
MAV_0714|M.avium_104 GRNIRINAIAPGPIDT---EANRTTTPKEMVDDIVKG-------LPLSRM
Mb0792|M.bovis_AF2122/97 GMKIRINAIAPGPIDT---EATRTVTPAELVKNMVQT-------IPLSRM
Rv0769|M.tuberculosis_H37Rv GMKIRINAIAPGPIDT---EATRTVTPAELVKNMVQT-------IPLSRM
MMAR_4927|M.marinum_M GMKIRINAIAPGPIDT---EATRTVTPGEFVKDMVKQ-------IPLSRM
MUL_0478|M.ulcerans_Agy99 GMKIRINAIAPGPIDT---EATRTVTPGEFVKDMVKQ-------IPLSRM
MAB_1219|M.abscessus_ATCC_1997 GQKIRINAIAPGPTDT---EATRGTVPEQFRSELVKN-------IPLSRM
MLBr_00862|M.leprae_Br4923 AVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRLRRY
. : :.:*.** ** ::.* .* : . : * *
Mflv_1601|M.gilvum_PYR-GCK GQPDDLVGMLLFLLS--DQAKWVTGQIFNVDGG-----QIIR--------
Mvan_5156|M.vanbaalenii_PYR-1 GEPEDLVGMLLFLLS--DQAKWITGQIFNVDGG-----QIIRS-------
TH_3037|M.thermoresistible__bu GEPEDLVGMCLFLLS--DQAKWITGQIFNVDGG-----QIIR--------
MSMEG_5858|M.smegmatis_MC2_155 GQPEDLVGMCLFLLS--DQARWITGQIFNVDGG-----QVIRS-------
MAV_0714|M.avium_104 GTPDDLVGMCLFLLS--DEASWITGQIFNVDGG-----QIIRS-------
Mb0792|M.bovis_AF2122/97 GTPEDLVGMCLFLLS--DSASWITGQIFNVDGG-----QIIRS-------
Rv0769|M.tuberculosis_H37Rv GTPEDLVGMCLFLLS--DSASWITGQIFNVDGG-----QIIRS-------
MMAR_4927|M.marinum_M GTPEDLVGMCLFLLG--DQASWITGQIFNVDGG-----QIIRS-------
MUL_0478|M.ulcerans_Agy99 GTPEDLVGMCLFLLG--DQASRITGQIFNVDGG-----QIIRS-------
MAB_1219|M.abscessus_ATCC_1997 GTVDDMVGMCLFLLS--DKASWITGQVFNVDGG-----QIIRS-------
MLBr_00862|M.leprae_Br4923 G-PDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTARIRVI
* :.:.. * : . :: : : : * *::*