For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADGTTLQRLSLRQLIWACLGVMAAAFVVVSAAAVAGQVGVARAVDELSNEVVPIQGEVEVLRRAFTDRE AGQRSYLLTGNPASLEPFDAGAVTADRMLTHLREELAGDPTGSALLDRIAAAAAAWTTDAAEPQIAARRS GPVPADEQASMTLESGRVFDRLRTELRALAAHTDAMTEAQLMQIQSVQRTANIVQAVGAVMLVLSVIGAV IAVRCLLTRPVGMLVDRVKRVADGDYDRPIIRPGLRETAEIADAVEQMRLSLRASTDRLIDSELRDEQAR IAAELNDRVIQRVFGLGLGLTAASARRDPDLQPFIDETDVIIRDLREVIFNLDHAVSPLGRPSRIRSAII DVVEGSVSALGFTPTLEIAGPVDDTPIPPELHAAVLAVVRESLSNVVRHSRATAAGVTIEVTGDKLRITV RDNGVGLASDNMMGRGRRNIASQAEQFGGTATVSTADSGTGAVVEWAVPLQPVGHLDAVTTSAR
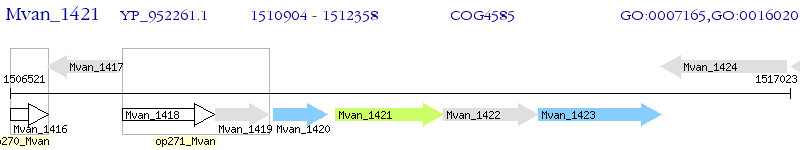
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1421 | - | - | 100% (484) | ATPase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1413 | - | 8e-23 | 38.00% (200) | GAF sensor signal transduction histidine kinase |
| M. vanbaalenii PYR-1 | Mvan_1395 | - | 3e-21 | 32.02% (228) | GAF sensor signal transduction histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3156c | devS | 2e-25 | 27.14% (420) | two component sensor histidine kinase DEVS |
| M. gilvum PYR-GCK | Mflv_3844 | - | 4e-22 | 31.60% (269) | GAF sensor signal transduction histidine kinase |
| M. tuberculosis H37Rv | Rv3132c | devS | 4e-25 | 27.14% (420) | two component sensor histidine kinase DEVS |
| M. leprae Br4923 | MLBr_00774 | mtrB | 3e-06 | 20.45% (313) | putative two-component system sensor kinase |
| M. abscessus ATCC 19977 | MAB_3890c | - | 3e-22 | 33.94% (221) | histidine kinase response regulator |
| M. marinum M | MMAR_1517 | devS | 9e-23 | 27.78% (378) | two-component sensor histidine kinase DevS |
| M. avium 104 | MAV_2508 | - | 5e-23 | 33.48% (233) | PAS domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3941 | - | 6e-24 | 34.10% (217) | GAF family protein |
| M. thermoresistible (build 8) | TH_0388 | - | 4e-20 | 33.06% (245) | TWO COMPONENT SENSOR HISTIDINE KINASE DEVS |
| M. ulcerans Agy99 | MUL_2422 | devS | 6e-23 | 27.87% (409) | two component sensor histidine kinase DevS |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3156c|M.bovis_AF2122/97 -------MTTGGLVDENDGAAMRPLRHTLSQLRLHELLVEVQDRVEQIVE
Rv3132c|M.tuberculosis_H37Rv -------MTTGGLVDENDGAAMRPLRHTLSQLRLHELLVEVQDRVEQIVE
MMAR_1517|M.marinum_M MTDEPHPQDAGPALGVPSGDGLRPLRLTLSQLRLRELLVEVQERVEQIVE
MUL_2422|M.ulcerans_Agy99 MTDEPHPQDAGPSLGVPSGDGLRPLRLTLSQLRLRELLVEVQERVEQIVE
Mflv_3844|M.gilvum_PYR-GCK ----MHDGAVADADDHRARDGARPVRTTLSQLRLRELLTEVQDRVEQIIR
MAB_3890c|M.abscessus_ATCC_199 -------------MVEDVGAQPNRVTAELAHLQLRELLAEVQGRIEQIVD
TH_0388|M.thermoresistible__bu --------------------------------------------------
MSMEG_3941|M.smegmatis_MC2_155 ---------------------------------------MGQSHTDDSPE
MAV_2508|M.avium_104 ----------------------------------------------MTSP
Mvan_1421|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00774|M.leprae_Br4923 -------------------------------------MIFSSRRRIRGRW
Mb3156c|M.bovis_AF2122/97 G-RDRLDGLVEAMLVVTAGLDLEATLRAIVHSATSLVDARYGAMEVHDRQ
Rv3132c|M.tuberculosis_H37Rv G-RDRLDGLVEAMLVVTAGLDLEATLRAIVHSATSLVDARYGAMEVHDRQ
MMAR_1517|M.marinum_M G-RDRLDGLVEAMLVVTSGLELNATLRTIVHSATNLVDARYCALEVHDRD
MUL_2422|M.ulcerans_Agy99 G-RDRLDGLVEAMLVVTSGLELDATLRTIVHSATNLVDARYCALEVHDRD
Mflv_3844|M.gilvum_PYR-GCK G-RDRLDGLVEAMLAVTSGLELDETLRTIVHTAIELVDAQYGALGVRGHD
MAB_3890c|M.abscessus_ATCC_199 GTRGRMDALLDAVMAVSSGLELDATLREIVCAASELVSARYGALGVVGSD
TH_0388|M.thermoresistible__bu --------------------------------------------------
MSMEG_3941|M.smegmatis_MC2_155 AVEVRMAALLQDMASLDAGLEPESTLHGILDAIMRVSGACYGAAGLCAPD
MAV_2508|M.avium_104 GADRYLSAAFRNKSLPERLRDLEAMVEAITDCAIIQLDAN----------
Mvan_1421|M.vanbaalenii_PYR-1 --------------MADGTTLQRLSLRQLIWACLGVMAAAFVVVSAAAVA
MLBr_00774|M.leprae_Br4923 GRSGPMMRGMGALTRVVGVVWRRSLQLRVVALTFGLSLAVILALGFVLTS
Mb3156c|M.bovis_AF2122/97 HRVLHFVYEGIDEETVRRIGHLPKGLGVIGLLIEDPKPLRLDDVSAHPAS
Rv3132c|M.tuberculosis_H37Rv HRVLHFVYEGIDEETVRRIGHLPKGLGVIGLLIEDPKPLRLDDVSAHPAS
MMAR_1517|M.marinum_M KRVLQFVYEGIDEDTVARIGHLPEGLGVIGLLIDEPKPLRLEDISQHPAS
MUL_2422|M.ulcerans_Agy99 KRVLQFVYEGIDEDTVARIGHLPEGLGIIGLLIDEPKPLRLEDISQHPAS
Mflv_3844|M.gilvum_PYR-GCK QELVEFIYEGIDEPMRERIGHLPEGRGVLGVLINDPKPIRLEDISRHADS
MAB_3890c|M.abscessus_ATCC_199 EKLAEFVYEGIDEATRDLIGPLPTGHGVLGVVIDEAKPLRLRDIAAHPAS
TH_0388|M.thermoresistible__bu --------------------------------------------------
MSMEG_3941|M.smegmatis_MC2_155 GTPATFLQAG---ALAQAVPDSPD-RDMLGFLRNRTGPLRIDGLARSAAE
MAV_2508|M.avium_104 GDVARWCPGAEAMTGYSAAEALGRPAALLYTSEDRAAGLAERELAAARES
Mvan_1421|M.vanbaalenii_PYR-1 GQVG--VARAVDELSNEVVPIQGEVEVLRRAFTDREAGQRSYLLTGNPAS
MLBr_00774|M.leprae_Br4923 QLTSRVLDVKVRVAIEQIERARTTVTGIVNGEETRSLDSSLQLARNTLTS
Mb3156c|M.bovis_AF2122/97 IGFPPYHPPMRTFLGVPVRVRDESFGTLYLTDKTNGQPFSDDDEVLVQAL
Rv3132c|M.tuberculosis_H37Rv IGFPPYHPPMRTFLGVPVRVRDESFGTLYLTDKTNGQPFSDDDEVLVQAL
MMAR_1517|M.marinum_M IGFPSHHPPMRTFLGVPVRVRDVSLGTLYLTDKTNGQPFSDDDELLVQAL
MUL_2422|M.ulcerans_Agy99 IGFPSHHPPMRTFLGVPVRVRDVSLGTLYLTDKTNGQPFSDDDELLVQAL
Mflv_3844|M.gilvum_PYR-GCK VGFPPNHPPMRTFLGVPVRIRDEVFGNLYLTEKSDGQPFSEDDEVLVQAL
MAB_3890c|M.abscessus_ATCC_199 VGFPANHPPMRTFLGVPVQVRGEVFGRLYLTEKLDGQEFTEDDEVVVRAL
TH_0388|M.thermoresistible__bu --------------------------------------------------
MSMEG_3941|M.smegmatis_MC2_155 VGLPELLPPMRAVLGVPLVSHGEVLGSLYVADSRAGFEFSDAHVGVVEVL
MAV_2508|M.avium_104 G-------------------RCEFEG---WRARKNGQRFRAG--VALSVF
Mvan_1421|M.vanbaalenii_PYR-1 LEP---------------FDAGAVTADRMLTHLREELAGDPTGSALLDRI
MLBr_00774|M.leprae_Br4923 KTDPTSGAGLVGAFDAVLIVPGDGPRTATTAGPVDQVPNSLRGFIKAGQA
Mb3156c|M.bovis_AF2122/97 AAAAGIAVANARLYQQAKARQSWIEATRDIATELLSGTE---PATVFRLV
Rv3132c|M.tuberculosis_H37Rv AAAAGIAVANARLYQQAKARQSWIEATRDIATELLSGTE---PATVFRLV
MMAR_1517|M.marinum_M AAAAGIAIANARLYEQAKARQAWIGATRDIATELLSGTE---PASVFRLV
MUL_2422|M.ulcerans_Agy99 AAAAGIAIANARLYEQAKARQAWIGATRDIATELLSGTE---PASVFRLV
Mflv_3844|M.gilvum_PYR-GCK AAAAGIAIDNARLYAQSQARQSWIEATRDIGTELLSGID---PAGVFRRV
MAB_3890c|M.abscessus_ATCC_199 AGAAGIAIENARLYQHARRREQWQRATADITAELLGGVD---PGKALHLV
TH_0388|M.thermoresistible__bu --------------------------------------------------
MSMEG_3941|M.smegmatis_MC2_155 ATVAADVVHSVWHLAKVRTVARWVSASREITMALLGMSGG--EDAPLRLI
MAV_2508|M.avium_104 TDDAGSAIG---FTTVMRDVTAEHQRAETMFHALLESAP----DAMVIVG
Mvan_1421|M.vanbaalenii_PYR-1 AAAAAAWTTDAAEPQIAARRSGPVPADEQASMTLESGRVFDRLRTELRAL
MLBr_00774|M.leprae_Br4923 AYQYATVHTEGFSGPALIIGTPTSSQVTNLELYLIFPLK----NEQATVT
Mb3156c|M.bovis_AF2122/97 AAEALKLTAADAALVAVPVDEDMPAADVGELLVIETVGSAVASTVGRTIP
Rv3132c|M.tuberculosis_H37Rv AAEALKLTAADAALVAVPVDEDMPAADVGELLVIETVGSAVASIVGRTIP
MMAR_1517|M.marinum_M AEEALKLTSADAALVAVPVDEDVPASEVAQLLVIETVGNAVAAAEGCTIA
MUL_2422|M.ulcerans_Agy99 AEEALKLTSADAALVAIPVDEDVPASEVAQLLVIETVGNAVAAAEGCTIA
Mflv_3844|M.gilvum_PYR-GCK ADRAQKLSGAAATLVAVPDDPDLPVGEVAELTVAASAGETIAAAH-DPIP
MAB_3890c|M.abscessus_ATCC_199 ASSAALLAEADFAFIALPAGDEE---DTAELVVAVCEGKGAEVLNDKLIP
TH_0388|M.thermoresistible__bu --------------------------------------------------
MSMEG_3941|M.smegmatis_MC2_155 VGRAAELTVASQAAILVPSEADRAPEEVRSLTVAVAAGARADEVEGQQVP
MAV_2508|M.avium_104 PDGRIVLANAQADQMFGYPREELIGREVEILIPPRHRGSHERYRTGFFAA
Mvan_1421|M.vanbaalenii_PYR-1 AAHTDAMTEAQLMQIQSVQRTANIVQAVGAVMLVLSVIGAVIAVRCLLTR
MLBr_00774|M.leprae_Br4923 LVRGTMATGGMVLLVLLSGIALLVSRQVVVPVRSASRIAERFAEGHLSER
Mb3156c|M.bovis_AF2122/97 VAGAVLREVFVNGIPRRVDRVDLEGLD-ELADAGPALLLPLRARGTVAGV
Rv3132c|M.tuberculosis_H37Rv VAGAVLREVFVNGIPRRVDRVDLEGLD-ELADAGPALLLPLRARGTVAGV
MMAR_1517|M.marinum_M VAGTALAEVLLDSAPRQVDKIAVEGVD-ELGDTGPALVLPLRATDAVAGV
MUL_2422|M.ulcerans_Agy99 VAGTALAEVLLDSAPRQVDKIAVEDVD-ELGDMGPALVLPLRATDAVAGV
Mflv_3844|M.gilvum_PYR-GCK VAGTAIGDAFVHRTPGMTDRADIG----IGATVGPALLLPLRAPDAVPGV
MAB_3890c|M.abscessus_ATCC_199 IDGSTSGSTFIDHEPRNVGRLAVNLAEGTDLAFGPALVLPLGGGESTAGV
TH_0388|M.thermoresistible__bu -------------------------------------------------V
MSMEG_3941|M.smegmatis_MC2_155 VSGSTSGRVFRTGESTMTDGFRYPITSFTDQGRRPAIVVPLRSATNNLGV
MAV_2508|M.avium_104 PAARRMGAGLELWGMR------------------------------CDGT
Mvan_1421|M.vanbaalenii_PYR-1 PVGMLVDR-----------------------------------------V
MLBr_00774|M.leprae_Br4923 MPVRGEDDMARLAVSFNDMAESLSRQITQLEEFGNLQRRFTSDVSHELRT
.
Mb3156c|M.bovis_AF2122/97 VVVLSQGGPGAFTDEQLEMMAAFADQAALA-WQLATSQRRMRELDVLTDR
Rv3132c|M.tuberculosis_H37Rv VVVLSQGGPGAFTDEQLEMMAAFADQAALA-WQLATSQRRMRELDVLTDR
MMAR_1517|M.marinum_M VVLLHPSSPESVTEEQLEMMAAFADQAALA-LQLATSQRRMRELDVMTER
MUL_2422|M.ulcerans_Agy99 VVLLHPSSPESVTEEQLEMMAAFADQAALA-LQLATSQRRMRELDVVTER
Mflv_3844|M.gilvum_PYR-GCK LIVLRSVGAQPFRSDELDMLAAFADQAAQA-WQLASTQHRLRELGILTDR
MAB_3890c|M.abscessus_ATCC_199 LILLRTVGAIAFDEEQLDMAASFAGQAALA-LEQAEVRLAKHELDVLADR
TH_0388|M.thermoresistible__bu LVVCRAEEGRPFTDEQLEILVTFADQAALA-WQLAATQRRMRELDVLSDR
MSMEG_3941|M.smegmatis_MC2_155 IAVARDADDPPFDVDEMAIMEDFAHHAAVA-LTLCRANEQARQLTVLSDR
MAV_2508|M.avium_104 VFPVDVSLSPLQTEQGVMVSAAIRDITEQL-AVQAELTETRAQAEVLAER
Mvan_1421|M.vanbaalenii_PYR-1 KRVADGDYDRPIIRPGLRETAEIADAVEQMRLSLRASTDRLIDSELRDEQ
MLBr_00774|M.leprae_Br4923 PLTTVRMAADLIYDHSSDLDPTLRRSTELMVSELDRFETLLNDLLEISRH
: . : :
Mb3156c|M.bovis_AF2122/97 DRIARDLHDHVIQRLFAIGLALQGAVPHERNPEVQQRLSDVVDDLQDVIQ
Rv3132c|M.tuberculosis_H37Rv DRIARDLHDHVIQRLFAIGLALQGAVPHERNPEVQQRLSDVVDDLQDVIQ
MMAR_1517|M.marinum_M DRIARDLHDHVIQRLFAIGLALQGSVPRTREPKVQQRLSEVIDDLQGVIQ
MUL_2422|M.ulcerans_Agy99 DRIARDLHDHVIQRLFAIGLALQGSVPRTREPKVQQRLSEVIDDLQGVIQ
Mflv_3844|M.gilvum_PYR-GCK DRIARDLHDHVIQRLFAVGLSLQGSIARARSEEVQQRLSANVDDLQSVIQ
MAB_3890c|M.abscessus_ATCC_199 DRIARDLHDHVIQRLFAVGLAMQSTHRRSVDPTVAARLADHIDQLHEVIQ
TH_0388|M.thermoresistible__bu DRIARDLHDHVIQRLFAVGLTLQGTIPRARSTEVQQRLSECIDDLQAVIG
MSMEG_3941|M.smegmatis_MC2_155 ERIARDLHDHVIQRLFLAGMDLQGTIARTKSDLLKERLSRTVDDLQSIID
MAV_2508|M.avium_104 DRIAGDLQDHAIQRVFAVGLALQGTIPRARSADVQQRLNAAVDDLHAVVQ
Mvan_1421|M.vanbaalenii_PYR-1 ARIAAELNDRVIQRVFGLGLGLTAASARRDPD-----LQPFIDETDVIIR
MLBr_00774|M.leprae_Br4923 DAGVAELSVEAVDLRVMVNNALGNVGHLAEEAGIELLVDMPVDEVIAEVD
. :* ..:: . . : : :*: :
Mb3156c|M.bovis_AF2122/97 EIRTTIYDLHGASQGI---TRLRQRIDAAVAQFA-DSGLRTSVQFVGPLS
Rv3132c|M.tuberculosis_H37Rv EIRTTIYDLHGASQGI---TRLRQRIDAAVAQFA-DSGLRTSVQFVGPLS
MMAR_1517|M.marinum_M EIRTTIFDLHGSSQGV---TRLRQRIDAAVAAFG-SSGLRTTVQFIGPLS
MUL_2422|M.ulcerans_Agy99 EIRTTIFDLHGSSQGV---TRLRQRIDAAVAAFG-SSGLRTTVQFIGPLS
Mflv_3844|M.gilvum_PYR-GCK EIRTAIFDLHGGSSAV---TRLRQRLDAAVAAFP-ESGVRTSMQCAGPLS
MAB_3890c|M.abscessus_ATCC_199 EIRTAIFDLHTNDE------SLRATLRSTINELTAETGLRVSVRMSGPLD
TH_0388|M.thermoresistible__bu EIRTTIFDLHGGPATG---SQIRQRIGEAANQFS-DAGVRTRVQFAGPLS
MSMEG_3941|M.smegmatis_MC2_155 EIRTAIFDLQSPAASA---MSFRQRIQAAVAAVTDNSPLATTVRISGPIV
MAV_2508|M.avium_104 DFRTAIFDLRHTKTDV---PGLRQRFDEVIGRLA--EGLAATVQYKGPLS
Mvan_1421|M.vanbaalenii_PYR-1 DLREVIFNLDHAVSPLGRPSRIRSAIIDVVEGSVSALGFTPTLEIAGPVD
MLBr_00774|M.leprae_Br4923 ARRVERILRNLIANAIDHSEHKPVRIRMAADEDTVAVTVRDYGIGLRPGE
* : . . *
Mb3156c|M.bovis_AF2122/97 --VVDSALADQAEAVVREAVSN----AVRHAKASTLTVRVKVD-DDLCIE
Rv3132c|M.tuberculosis_H37Rv --VVDSALADQAEAVVREAVSN----AVRHAKASTLTVRVKVD-DDLCIE
MMAR_1517|M.marinum_M --VVDNVLADHAEAVVREAVSN----AVRHAQATTLTVRVKVD-DDLCIE
MUL_2422|M.ulcerans_Agy99 --VVDNVLADHAEAVVREAVSN----AVRHAQATTLTVRVKVD-DDLCIE
Mflv_3844|M.gilvum_PYR-GCK --VVDAGLADHAEAVVREAVSN----AVRHAEASTVAVTVTVE-DDLAIT
MAB_3890c|M.abscessus_ATCC_199 --VVPASTSGDAEAVVREAVSN----VVRHARARELSVTVSVD-DELVIE
TH_0388|M.thermoresistible__bu --VISAVLADHAEAVVREAVSN----AVRHGGATDLSVDVSVD-DDLTIE
MSMEG_3941|M.smegmatis_MC2_155 --AVDPALADDAEAVIVEAISN----AVRHSGATSVTISIEVS-DRLDIE
MAV_2508|M.avium_104 --VVEGELADQAEAVVAEAIGN----AVRHTAATKLTIAVEVA-DEVSIE
Mvan_1421|M.vanbaalenii_PYR-1 DTPIPPELHAAVLAVVRESLSN----VVRHSRATAAGVTIEVTGDKLRIT
MLBr_00774|M.leprae_Br4923 EKLVFSRFWRSDPSRVRRSGGTGLGLAISIEDARLHQGRLEAWGEPGQGA
: : : .: .. .: * : . :
Mb3156c|M.bovis_AF2122/97 VTDNGRGLPD-EFTGSGLTNLRQRAEQAGGEFTLASVPGASGTVLRWSAP
Rv3132c|M.tuberculosis_H37Rv VTDNGRGLPD-EFTGSGLTNLRQRAEQAGGEFTLASVPGASGTVLRWSAP
MMAR_1517|M.marinum_M VSDNGRGMPK-TYTSSGLTNLRRRAQEAGGQFTIESPAAGGGTLLRWSAP
MUL_2422|M.ulcerans_Agy99 VSDNGRGMPK-TYTSSGLTNLRRRAQEAGGQFTIESPAAGGGTLLRWSAP
Mflv_3844|M.gilvum_PYR-GCK VTDDGRGIAA-DVTGSGLANLARRAQDVGGSFSVR-RGAQVGTTLTWRAP
MAB_3890c|M.abscessus_ATCC_199 VVDDGIGIPD-VVARSGLSGLANRAAQSGGTFSVSAMDTG-GTRLVWSAP
TH_0388|M.thermoresistible__bu VTDNGRGFPEGEVTESGLANLRHRARHAGGTLEVHNRPDG-GARLHWSVP
MSMEG_3941|M.smegmatis_MC2_155 VVDNGRGIPANTARCSGLANLKARAQQAGGSCQITSSAGS-GTHVRWSAP
MAV_2508|M.avium_104 VIDNGKGLPD-DVSEAGLKTLRRRAERVGGTLTVGAAAGG-GTRLRWVAP
Mvan_1421|M.vanbaalenii_PYR-1 VRDNGVGLASDNMMGRGRRNIASQAEQFGGTATVSTADSGTGAVVEWAVP
MLBr_00774|M.leprae_Br4923 CFRLTLPLVRGHKVTTSPLPMKPILQPSPQASTAGQQHGTQRQRLREHAE
: . : : .
Mb3156c|M.bovis_AF2122/97 LSQ------------
Rv3132c|M.tuberculosis_H37Rv LSQ------------
MMAR_1517|M.marinum_M LIQ------------
MUL_2422|M.ulcerans_Agy99 LIQ------------
Mflv_3844|M.gilvum_PYR-GCK LP-------------
MAB_3890c|M.abscessus_ATCC_199 LP-------------
TH_0388|M.thermoresistible__bu LT-------------
MSMEG_3941|M.smegmatis_MC2_155 LSGA-----------
MAV_2508|M.avium_104 LP-------------
Mvan_1421|M.vanbaalenii_PYR-1 LQPVGHLDAVTTSAR
MLBr_00774|M.leprae_Br4923 RSR------------