For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLAMVYDGPGRRSWRSVPEPVIQQGTDAIVRVDAVTICGTDLHILKGDVPEVEPGRVLGHEAVGTVTEVG PAVQTLSVGDRVLVSCISACGACRYCRQGQYGQCLGGGGWILGHLIDGTQAEYVRVPFADNSTHKVPEGV SDEQMIVLADILPTSYEVGVLAGAVSPGDVVAIVGAGPIGLAAVLTAKLYSPSHIVVIDVADARLDAARR FGADTVVNSSTRSARDVIDELTGGLGADVAMEAVGLPETFEESVALVRPGGHVANIGVHGAPATLHLEQI WIKNLTITTGLVDTHSTPTLIRLVANHQLDTAAMITHRFPMDEFDTAYDVFGDAAHSGALKVLLTASP
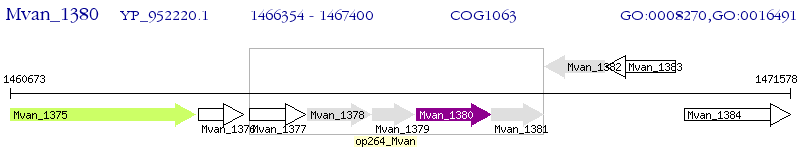
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1380 | - | - | 100% (348) | alcohol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4901 | - | e-143 | 69.57% (345) | alcohol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1841 | - | 5e-46 | 35.76% (330) | alcohol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0167c | adhE1 | 7e-30 | 27.86% (359) | putative zinc-type alcohol dehydrogenase (e subunit) adhE |
| M. gilvum PYR-GCK | Mflv_4997 | - | 1e-175 | 86.74% (347) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv1895 | - | 2e-48 | 34.24% (330) | dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 3e-29 | 33.99% (253) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_2084 | - | 6e-30 | 28.48% (330) | zinc-type alcohol dehydrogenase AdhD |
| M. marinum M | MMAR_2790 | - | 1e-48 | 34.68% (346) | dehydrogenase |
| M. avium 104 | MAV_2804 | - | 7e-50 | 35.55% (346) | zinc-binding dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0671 | - | 4e-51 | 34.51% (368) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. thermoresistible (build 8) | TH_2319 | - | 3e-37 | 38.08% (239) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. ulcerans Agy99 | MUL_2962 | - | 1e-46 | 33.82% (346) | dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1380|M.vanbaalenii_PYR-1 ---MLAMVYDGP-------------------GRRSWRSVPEPVIQQGTDA
Mflv_4997|M.gilvum_PYR-GCK ---MQAMVYTGP-------------------GQRSWQTVPDPVLVDATDA
MMAR_2790|M.marinum_M ---MRTVVIDGP-------------------GSIRVDNRPDPALPGSDGV
MUL_2962|M.ulcerans_Agy99 ---MRTVVIDGP-------------------GSIRVDNRPDPALPGSDGV
Rv1895|M.tuberculosis_H37Rv ---MRAVVIDGA-------------------GSVRVNTQPDPALPGPDGV
MAV_2804|M.avium_104 ---MRTVVVDGP-------------------RSIRVDTRPDPVLPGPDGA
MSMEG_0671|M.smegmatis_MC2_155 ---MRAMVYRGP-------------------YKIRLEDKPIPAIEHPNDA
Mb0167c|M.bovis_AF2122/97 MPAVQPWLYSNMPAIRGAVLDQIGVPRPYWRSKPISVVELHLDPPDRGEV
MAB_2084|M.abscessus_ATCC_1997 MKTRAAVLWG--------------------LEQKWEVEEVDLDPPGPGEV
MLBr_01784|M.leprae_Br4923 MSQTVRGVISRK------------------KDEPVELVDIVIPDPGPGEA
TH_2319|M.thermoresistible__bu ------VTWHGR-------------------RKVSVDTVPDPAIMEPTDA
.
Mvan_1380|M.vanbaalenii_PYR-1 IVRVDAVTICGTDLHILKGDVPEVEPGRVLGHEAVGTVTEVGPAVQTLSV
Mflv_4997|M.gilvum_PYR-GCK IVRVDTVTICGTDLHILKGDVPEVESGRILGHEAVGTVTAIGSAVQTLSV
MMAR_2790|M.marinum_M IVAVSAAGICGSDLHFYEGEYPLAEP-VALGHEAVGTVVEKGPDVHTVEV
MUL_2962|M.ulcerans_Agy99 IVVVSAAGICGSDLHFYEGEYPLAEP-VALGHEAVGTVVEKGPDVHTVEV
Rv1895|M.tuberculosis_H37Rv VVAVTAAGICGSDLHFYEGEYPFTEP-VALGHEAVGTIVEAGPQVRTVGV
MAV_2804|M.avium_104 IVEVSAAGICGSDLHFYEADFPMPDP-IALGHEAIGTVVEAGPQVRNVKV
MSMEG_0671|M.smegmatis_MC2_155 IVRVTKAAICGSDLHLYHGMMPDTRVGTTFGHEFIGVVEKVGPSVQNLSV
Mb0167c|M.bovis_AF2122/97 LVRIEAAGVCHSDLSVVDGTRVRPVP-ILLGHEAAGIVEQVGDGVDGVAV
MAB_2084|M.abscessus_ATCC_1997 LVRLAATGLCHSDEHLVTGDLPIPLP-VVGGHEGAGTVVECGEGVEDLSE
MLBr_01784|M.leprae_Br4923 VVDVTACGVCHTDLTYREGGINDRYP-FLLGHEAAGTVEVVGPGVTAVEP
TH_2319|M.thermoresistible__bu IIRVTSTNICGSDLHLYEVLSAFMSPGDILGHEAMGVVEEVGPEVGALQA
:: : :* :* *** * : * * :
Mvan_1380|M.vanbaalenii_PYR-1 GDRVLVSCISACGACRYCRQGQYGQCLGGGG-------WILG--------
Mflv_4997|M.gilvum_PYR-GCK GDRVLVSCISSCGACRYCRQGHYGQCLGGGG-------WILG--------
MMAR_2790|M.marinum_M GDQVMVSSVAGCGACAGCATRDPAMCVSGPQ--------IFG--------
MUL_2962|M.ulcerans_Agy99 GDQVMVSSVAGCGACAGCATRDPAMCVSGPQ--------IFG--------
Rv1895|M.tuberculosis_H37Rv GDLVMVSSVAGCGVCPGCETHDPVMCFSGPM--------IFG--------
MAV_2804|M.avium_104 GDQVMVSSVAGCGSCAGCATRDPVLCHSGFQ--------IFG--------
MSMEG_0671|M.smegmatis_MC2_155 GDRVMVPFNIFCGSCYFCARGLYSNCHNVNPNATAV-GGIYGY------S
Mb0167c|M.bovis_AF2122/97 GQRVVLVFLPRCGQCAACATDGRTPCEPGSAANKAG-TLLGGGIRLSRGG
MAB_2084|M.abscessus_ATCC_1997 GDSVILTFLPSCGRCSYCARGMGNLCELG-AALMMG-PQIDGTYRFHARG
MLBr_01784|M.leprae_Br4923 GDFVILNWRAVCGQCRACKRGRPSYCFDTFNAQQKM-TLIDG--------
TH_2319|M.thermoresistible__bu GDRVVIPFNISCGTCWMCAQGLQSQCETTQNRDQGTGAALFG------YS
*: *:: ** * * * : *
Mvan_1380|M.vanbaalenii_PYR-1 ---HLIDGTQAEYVRVPFADNSTHKVPEGVSDEQMIVLADILPTSYEVGV
Mflv_4997|M.gilvum_PYR-GCK ---HLIDGTQAEYVRVPFADNSTHKVPAGVSDEQMIVLADILPTSYEVGV
MMAR_2790|M.marinum_M --SGALGGAQADLLAVPAADFQVLKIPAAITTEQALLLTDNLATGWAAAR
MUL_2962|M.ulcerans_Agy99 --SGALGGAQADLLAVPATDFQVLKIPAAITTEQALLLTDNLATGWAAAR
Rv1895|M.tuberculosis_H37Rv --AGVLGGAQADLLAVPAADFQVLKIPEGITTEQALLLTDNLATGWAAAQ
MAV_2804|M.avium_104 --GGVLGGAQADLLAVPAADFQLLKIPEGISTEQALLLTDNLATGWAAAQ
MSMEG_0671|M.smegmatis_MC2_155 HTCGGYDGGQAEFVRVPFADVGPSIIPDRLDDEDALLLTDALATGYFGAQ
Mb0167c|M.bovis_AF2122/97 RPVYHHLGVSGFATHVVVNRASVVPVPHEVPPTVAALLGCAVLTGGGAVL
MAB_2084|M.abscessus_ATCC_1997 EDVGQMCLLGTFSEYTVVPQASVVKIDGGIPLDRAALIGCGVTTGYGSAV
MLBr_01784|M.leprae_Br4923 TELTPALGIGAFADKTLVHSGQCTKVDPAADPAVAGLLGCGVMAGLGAAI
TH_2319|M.thermoresistible__bu KLYGEVAGGQAEYLRVPQAQYTHITVPHDGPDDRYVYLSDVLPTAWQGVE
. : : : :.
Mvan_1380|M.vanbaalenii_PYR-1 LAGAVSPGDVVAIVGAGPIGLAAVLTAKLYSPSHIVVIDVADARLDAARR
Mflv_4997|M.gilvum_PYR-GCK LAGRVGPGDVVAIVGAGPIGLAAVLTAKLYSPSHIVVIDVADTRLEAAKK
MMAR_2790|M.marinum_M RA-DILFGATVAVIGLGAVGLCAVRSALAQGAATVFAVDRVQGRLQRASE
MUL_2962|M.ulcerans_Agy99 RA-DIPFGATVAVIGLGAVGLCAVRSALAQGAATVFAVDRVQGRLQRAAE
Rv1895|M.tuberculosis_H37Rv RA-DISFGSAVAVIGLGAVGLCALRSAFIHGAATVFAVDRVKGRLQRAAT
MAV_2804|M.avium_104 RA-DIPFGGTVAVIGLGAVGLCALRSALFQGAATVFAVDQVDGRLARAAR
MSMEG_0671|M.smegmatis_MC2_155 LG-NIAEGDTVVVFGAGPVGLFAAKSAWLMGAGRVIVIDHLEYRLEKART
Mb0167c|M.bovis_AF2122/97 NVGDPQPGQSVAVVGLGGVGMAAVLTALTYTDVRVVAVDQLPEKLSAAKA
MAB_2084|M.abscessus_ATCC_1997 RTGEVRAGDTVVVVGAGGIGMNAIQGARIAGALAIVAVDPVNFKREQASG
MLBr_01784|M.leprae_Br4923 NTAAVSRDDTVAVIGCGGVGDAAISGAALVGANRIIAVDIDDTKLEWART
TH_2319|M.thermoresistible__bu YA-AVPDGGTLVVLGLGPIGSMACRIAAHRGNCRVIGVDRVPERIEKVRP
. :.:.* * :* * * :. :* : .
Mvan_1380|M.vanbaalenii_PYR-1 FGADTVVNSSTRSARDVIDELTGG-------------LGADVAMEAVGLP
Mflv_4997|M.gilvum_PYR-GCK LGADVTVNSRSRRPRDVIDELTGG-------------LGADVVMEAVGLP
MMAR_2790|M.marinum_M WG-ATAVASP--AAEAIVAATGGR--------------GADSVIDAVGTD
MUL_2962|M.ulcerans_Agy99 WG-ATAVASP--AAEAIVAATSGR--------------GADSVIDAVDTD
Rv1895|M.tuberculosis_H37Rv WG-ATPIPSP--AAETILAATRGR--------------GADSVIDAVGTD
MAV_2804|M.avium_104 WG-ATPLKAP--ALEAILAATGGR--------------GADAVIDAVATD
MSMEG_0671|M.smegmatis_MC2_155 FAHAETINFV--DCDDIVVKMKRT----------TDHLGADVAIDAAGAE
Mb0167c|M.bovis_AF2122/97 LGA--------HEIYTPQQATAGG-------------VKAAVVVEAVGHP
MAB_2084|M.abscessus_ATCC_1997 FGATHTA-ASMDEAWSLVSDMTRG-------------KLADVCILTTDVA
MLBr_01784|M.leprae_Br4923 FGATHTVNALELEVVKTIQDLTSG-------------FGVDVVIDTVGRP
TH_2319|M.thermoresistible__bu YCADVLNVDTDDVEAAVREQTGGXXVQTPPDPRR-QSADCENHADQHGRP
Mvan_1380|M.vanbaalenii_PYR-1 ET---------------------FEESVALVRPG----------------
Mflv_4997|M.gilvum_PYR-GCK ET---------------------FEESVDLVRPC----------------
MMAR_2790|M.marinum_M AS---------------------MTDALNAVRAG----------------
MUL_2962|M.ulcerans_Agy99 AS---------------------MTDALNAVRAG----------------
Rv1895|M.tuberculosis_H37Rv AS---------------------MSDALNAVRPG----------------
MAV_2804|M.avium_104 AS---------------------LTEALNAVRPG----------------
MSMEG_0671|M.smegmatis_MC2_155 ADGNFLQHVTATKLKLQGGSPIALNWAIDSVRKG----------------
Mb0167c|M.bovis_AF2122/97 AA---------------------LHTAIGLTAPG--------------GR
MAB_2084|M.abscessus_ATCC_1997 EG---------------------SYTAEALALVGK------------RGR
MLBr_01784|M.leprae_Br4923 ET---------------------WKQAFYARDLA--------------GT
TH_2319|M.thermoresistible__bu EHP-------PEELHHQADDQRQRRHERDAATAGSPPLPSPWPDRTTTHR
Mvan_1380|M.vanbaalenii_PYR-1 GHVANIGVHG-APATLHLEQIWIKNLTIT---TGLVDTH-STPTLIRLVA
Mflv_4997|M.gilvum_PYR-GCK GHVANIGVHG-APATLHLERIWIKNLTIT---TGLVDTH-STPTLISLVA
MMAR_2790|M.marinum_M GTVSVVGVHDLQPFPLPALTCLLRSITLR---LTIAPVQRTWSELIPLLE
MUL_2962|M.ulcerans_Agy99 GTVSVVGVHDLQPFPLPALTCLLRSITLR---LTIAPVQRTWSELIPLLE
Rv1895|M.tuberculosis_H37Rv GTVSVVGVHDLQPFPVPALTCLLRSITLR---MTMAPVQRTWPELIPLLQ
MAV_2804|M.avium_104 GTVSVVGVHDMNPFPLNALGCLIRSITLR---MTTAPVQRTWPELIPLLQ
MSMEG_0671|M.smegmatis_MC2_155 GTVSVVGAYGPMFSAVKFGDAFNKGLTLR---MNQCPVKREWPRLLSHIK
Mb0167c|M.bovis_AF2122/97 TITVGLPPPD-VRISLSPLDFVTEGRSLIGSYLGSAVPSHDIPRFVSLWQ
MAB_2084|M.abscessus_ATCC_1997 VVVTAIGHPDESSISGSLLEMTLYEKQIRGALYGSSNAPHDIPRLVELYS
MLBr_01784|M.leprae_Br4923 VVLVGVPTPD-MRLDMPLLDFFSHGGSLKSSWYGDCLPERDFPTLVDLYL
TH_2319|M.thermoresistible__bu HTWRRLGGMELLRDVVVLLHIVGFAVTFGAWTAEAVARRFRTTRLMDYGL
: : . ::
Mvan_1380|M.vanbaalenii_PYR-1 NHQLDTAAMITHRFPMDEFDTAYDVFGDAAHSGALKVLLTASP-------
Mflv_4997|M.gilvum_PYR-GCK NGRLDAATMITHRYPMDEFDTAYQVFGDAANSGALKVVLTATS-------
MMAR_2790|M.marinum_M SGRLDVDGIFTTSLPLDEAAKGYSIAGSRSGN-DVKVLLKV---------
MUL_2962|M.ulcerans_Agy99 SGRLDVDGIFTTSLPLDEAAKGYSIAGSRSGN-DVKVLLKV---------
Rv1895|M.tuberculosis_H37Rv SGRLDVDGIFTTTLPLDEAAKGYATARARSGE-ELRFCLRPDSRDVLGAH
MAV_2804|M.avium_104 SGRLDVDGIFTTTMPLDEAAIGYATAGSRSGD-DVKILLKP---------
MSMEG_0671|M.smegmatis_MC2_155 NGYLKPNDIVTHRIPLEHIAEGYHIFSAKLDD-CIKPIIVPTAS------
Mb0167c|M.bovis_AF2122/97 SGRLPVESLVTSTIRLDDINEAMDHLADGIAVRQLISFTGDL--------
MAB_2084|M.abscessus_ATCC_1997 AGLLKLDELVTREYSLDQINEGYQDMRSGRNIRGLVRF------------
MLBr_01784|M.leprae_Br4923 QGRLPLGKFVSERIGLGDVEEAFHKIHGGKVLRSVVML------------
TH_2319|M.thermoresistible__bu LLSLITGLALAAPWPAGIVLNYPKIGLKLVLLVALGAVLGIGGAHQRRTG
* .: :
Mvan_1380|M.vanbaalenii_PYR-1 ----------------------------------
Mflv_4997|M.gilvum_PYR-GCK ----------------------------------
MMAR_2790|M.marinum_M ----------------------------------
MUL_2962|M.ulcerans_Agy99 ----------------------------------
Rv1895|M.tuberculosis_H37Rv ETVDLYVHVRRCQSVADLQLEGAADGVDGPSMLN
MAV_2804|M.avium_104 ----------------------------------
MSMEG_0671|M.smegmatis_MC2_155 ----------------------------------
Mb0167c|M.bovis_AF2122/97 ----------------------------------
MAB_2084|M.abscessus_ATCC_1997 ----------------------------------
MLBr_01784|M.leprae_Br4923 ----------------------------------
TH_2319|M.thermoresistible__bu NPVPRPLFVTAGVLSFAAAAVAVLW---------