For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGPFRFPSAWREFLAAEPVRVSAVLRLPLIGLIAVLVWIWEVNHWLPGLYAAILGSYAVVAVVWLLAVSR GPVPRWADWASSAVDVLVIVSLCVVSGGATAALLPVFFLLPISVAFQDRPALTAIIGSVTAVAYLTVWVF YSRRDDEVGLPNMVYTQFGFLVWMAVATTALSFVLAHRAVRLRNLQEVRVQLVSEAMQSDERHNREVAES LHDGPLQTLLAARLELDEARERNPDPALDAVYAALQETADGLRATVTELHPQVLAQLGLTAGIRELLRQL QSRHLLEVDAELEEVGEPQSQALLYRAARELLTNIVKHADATTVRVRLARRGDRVVLSVADDGAGFDPAI LGTSLGQGHIGLGSLIARFDAMGGSMRITSSDGQGTEVTATSPPEPA*
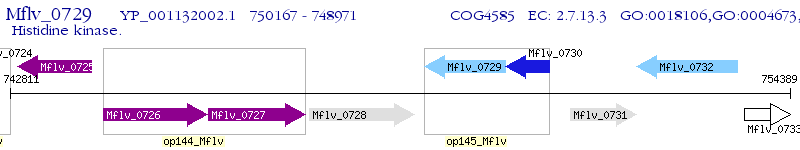
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0729 | - | - | 100% (398) | integral membrane sensor signal transduction histidine kinase |
| M. gilvum PYR-GCK | Mflv_5015 | - | 2e-22 | 32.46% (228) | GAF sensor signal transduction histidine kinase |
| M. gilvum PYR-GCK | Mflv_0967 | - | 2e-12 | 29.21% (202) | integral membrane sensor signal transduction histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0868 | - | 6e-41 | 31.79% (390) | two component sensor kinase |
| M. tuberculosis H37Rv | Rv0845 | - | 4e-41 | 31.79% (390) | two component sensor kinase |
| M. leprae Br4923 | MLBr_00774 | mtrB | 9e-05 | 23.96% (192) | putative two-component system sensor kinase |
| M. abscessus ATCC 19977 | MAB_0061 | - | 1e-104 | 50.78% (386) | two component sensor kinase |
| M. marinum M | MMAR_4781 | - | 7e-38 | 30.61% (392) | two-component sensor kinase |
| M. avium 104 | MAV_0874 | - | 4e-36 | 30.85% (376) | hypothetical protein MAV_0874 |
| M. smegmatis MC2 155 | MSMEG_0106 | - | 1e-148 | 68.25% (378) | sensory histidine kinase |
| M. thermoresistible (build 8) | TH_4203 | - | 4e-22 | 34.40% (218) | PUTATIVE sensor histidine kinase |
| M. ulcerans Agy99 | MUL_0350 | - | 1e-37 | 30.61% (392) | two component sensor kinase |
| M. vanbaalenii PYR-1 | Mvan_1357 | - | 8e-23 | 33.91% (233) | GAF sensor signal transduction histidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0729|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0106|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0061|M.abscessus_ATCC_1997 --------------------------------------------------
Mb0868|M.bovis_AF2122/97 --------------------------------------MPSYGNLGRLGG
Rv0845|M.tuberculosis_H37Rv --------------------------------------MPSYGNLGRLGG
MMAR_4781|M.marinum_M --------------------------------------------------
MUL_0350|M.ulcerans_Agy99 MMNSRQREADRRQFAPSCRRQDESAGQSLGQRRTERSPNPSCANYGQLVG
MAV_0874|M.avium_104 --------------------------------------------------
TH_4203|M.thermoresistible__bu -------------------------------------------------V
Mvan_1357|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00774|M.leprae_Br4923 -MIFSSRRRIRGRWGRSGPMMRGMGALTRVVGVVWRRSLQLRVVALTFGL
Mflv_0729|M.gilvum_PYR-GCK ---------MTGPFRFPSAWREFLAAEPVRVSAVLRLPLIGLI-------
MSMEG_0106|M.smegmatis_MC2_155 --------------MISGRIVEFFASQPVRVSAVLRLPLIGLI-------
MAB_0061|M.abscessus_ATCC_1997 -------------MTRLDNMADYFAAEPMRVSAWLRLPLIGLI-------
Mb0868|M.bovis_AF2122/97 -RHEYGVLVAMTSSAELDRVRWAHQLRSYRIASVLRIGVVGLM-------
Rv0845|M.tuberculosis_H37Rv -RHEYGVLVAMTSSAELDRVRWAHQLRSYRIASVLRIGVVGLM-------
MMAR_4781|M.marinum_M ---------MVTSDAELARVRRTHQLRAYRIGSVLRIGVVLFM-------
MUL_0350|M.ulcerans_Agy99 SCRLYGVPVVVTSDAELARVRRTHQLRAYRIGSVLRIGVVLFM-------
MAV_0874|M.avium_104 --------MAVAGDAELERVRAVHQMRSYRIGSVLRVGVVGLM-------
TH_4203|M.thermoresistible__bu TSGPHPDPVRDLSEQLLTADRELVLLRELIQAASRGPGVEPLA-------
Mvan_1357|M.vanbaalenii_PYR-1 -----------MSRPLLTADRELALLRELIQAASSGPGVEPLA-------
MLBr_00774|M.leprae_Br4923 SLAVILALGFVLTSQLTSRVLDVKVRVAIEQIERARTTVTGIVNGEETRS
: :
Mflv_0729|M.gilvum_PYR-GCK ----AVLVWIWEVNHWLPGLYAAILGSYAVVAVVWLLAVSR---------
MSMEG_0106|M.smegmatis_MC2_155 ----FVLVMVWEVDHWLPGLYAVILGLYAAAAVLWLVVVIR---------
MAB_0061|M.abscessus_ATCC_1997 ----VLLGSDPNIQMWHNGVYYGVLAAYTLSAVVWVGIAVR---------
Mb0868|M.bovis_AF2122/97 ----VAAMVVGTSRSEWP-QQIVLIGVYAVAALWALLLAYSASRRFFALR
Rv0845|M.tuberculosis_H37Rv ----VAAMVVGTSRSEWP-QQIVLIGVYAVAALWALLLAYSASRRFFALR
MMAR_4781|M.marinum_M ----VAAMLVGTPRREWG-AQTALICVYAFAALWALALAFRRSRPMVSLD
MUL_0350|M.ulcerans_Agy99 ----VAAMLVGTPRREWG-AQTALICVYAFAALWALALAFRRSRPMVSLD
MAV_0874|M.avium_104 ----VAAMIIGTARSEWP-QESVLILLYGIIALGAVALAFAPFRRWIGIG
TH_4203|M.thermoresistible__bu ----AAAARMITAATASDVCFVHVLDDTERSLTLAGATPPFD--------
Mvan_1357|M.vanbaalenii_PYR-1 ----AAAARMITAATDSDVCFVHVLDDTERSLTLAGATPPFD--------
MLBr_00774|M.leprae_Br4923 LDSSLQLARNTLTSKTDPTSGAGLVGAFDAVLIVPGDGPRTATTAGPVDQ
::
Mflv_0729|M.gilvum_PYR-GCK --GPVPRWADWASSAVDVLVIVSL-----CVVSGGATAALLPVFFLLPIS
MSMEG_0106|M.smegmatis_MC2_155 --GPMPRWAEWASTAVDGLVLLAL-----CAVSGGATAALLPVFFLLPIS
MAB_0061|M.abscessus_ATCC_1997 --GQVADWVAPAATSADIAAVVML-----CLASGAGNTELLPVFFLLPVS
Mb0868|M.bovis_AF2122/97 RFRSMGRLEPFAFTAVDVLILTGF-----QLLSTDGIYPLL-IMILLPVL
Rv0845|M.tuberculosis_H37Rv RFRSMGRLEPFAFTAVDVLILTGF-----QLLSTDGIYPLL-IMILLPVL
MMAR_4781|M.marinum_M RFANIARYEPLAFTAVDVLALTGF-----QLLSTDGIYSLL-IMTLLPVL
MUL_0350|M.ulcerans_Agy99 RFANIARYEPLAFTAVDVLALTGF-----QLLSTDGIYSLL-IMTLLPVL
MAV_0874|M.avium_104 RLAGVGRLEPFAFTVVDILALTIF-----QLLSTNGIYPLL-IMAMLPVL
TH_4203|M.thermoresistible__bu --KEVGKIRLPLGQGISGWVASHR-----EPVVITENKEADPRYLPFQSL
Mvan_1357|M.vanbaalenii_PYR-1 --SEVGKIRLPLGQGISGWVASNR-----EPVVIRRDKESDPRYLPFRSL
MLBr_00774|M.leprae_Br4923 VPNSLRGFIKAGQAAYQYATVHTEGFSGPALIIGTPTSSQVTNLELYLIF
: .
Mflv_0729|M.gilvum_PYR-GCK VAFQDRPALTAIIGSVTAVAYLTVWVFYSRRDDEVGLPNMVYTQFGFLVW
MSMEG_0106|M.smegmatis_MC2_155 VAFQDRPLLTAMLGTATATGYLGAWIFYSKRDDTVGLPNVVYMQVGFLLW
MAB_0061|M.abscessus_ATCC_1997 VAFQERPAITAILGIITAVAYFGILVYYIADGNLERIPGDEYVTWAALLW
Mb0868|M.bovis_AF2122/97 VGLDVSTRRAAVVLACTLVGFAVAVLGDPVMLRAIGWPETIFR-FALYAF
Rv0845|M.tuberculosis_H37Rv VGLDVSTRRAAVVLACTLVGFAVAVLGDPVMLRAIGWPETIFR-FALYAF
MMAR_4781|M.marinum_M VALDISSRRAAVVLALTLVGFALALAQDPVVIRMNGWPEAIFR-FGLYAF
MUL_0350|M.ulcerans_Agy99 VALDISSRRAAVVLALTLVGFGLALAQDPVVIRMNGWPEAIFR-FGLYAF
MAV_0874|M.avium_104 LGLDVSSRRAAVVLIFSMLGFAIAVLQDPVMSSSVRLPEAGFR-FLLYGF
TH_4203|M.thermoresistible__bu RGRDFTS-MVSVPMETDPGGLVGVLNVHTVERREFTRRD------VDLLL
Mvan_1357|M.vanbaalenii_PYR-1 RGRDFTS-MVSVPMETDPGGLVGVLNVHTVAERDFGDRD------VGLLL
MLBr_00774|M.leprae_Br4923 PLKNEQATVTLVRGTMATGGMVLLVLLSGIALLVSRQVVVPVRSASRIAE
: . . : .
Mflv_0729|M.gilvum_PYR-GCK MAVATTALSFVLAH----RAVRLRNLQEVRVQLVSEAMQSDER--HNREV
MSMEG_0106|M.smegmatis_MC2_155 LAVATTALSLVLVR----RSARVRALLDVRRRLVQESARADEL--RDAEL
MAB_0061|M.abscessus_ATCC_1997 LTVFTTGMCFVLKR----RSARVVQLVHIREQLVLESMRADER--RNREV
Mb0868|M.bovis_AF2122/97 LCATALMVVRIEER----HTRSVAGLSALREELLAQTMTASEV--LQRRI
Rv0845|M.tuberculosis_H37Rv LCATALMVVRIEER----HTRSVAGLSALRAELLAQTMTASEV--LQRRI
MMAR_4781|M.marinum_M LCATALMVVRSEER----HARSVATLSVIREELLAQTMTASEV--LQRRI
MUL_0350|M.ulcerans_Agy99 LCATALMVVRSEER----HARSVATLSVIREELLAQTMTASEV--LQRRI
MAV_0874|M.avium_104 LCCAAFLVVRIEER----HTRSVAGLSALREELLAQTMNASDV--SQRRI
TH_4203|M.thermoresistible__bu VIGRLIAGGLHQAR----LHRQLVARERAHEIFVEQVIEAQEL--ERRRL
Mvan_1357|M.vanbaalenii_PYR-1 VIGRLIAGAMHQAR----LHRQLVARERAHEHFVEQVIEAQEL--ERRRL
MLBr_00774|M.leprae_Br4923 RFAEGHLSERMPVRGEDDMARLAVSFNDMAESLSRQITQLEEFGNLQRRF
: : : .: ..
Mflv_0729|M.gilvum_PYR-GCK AESLHDGPLQTLLAARLELDEARERNP--------DPALDAVYAALQETA
MSMEG_0106|M.smegmatis_MC2_155 AEALHDGPLQTLLAARLELDELRERNP--------DPALDVVHAALQQTA
MAB_0061|M.abscessus_ATCC_1997 AERLHDGPLQNLLAARLRIEEVLERQS--------DPVLTAVHSSLRETA
Mb0868|M.bovis_AF2122/97 AEAIHDGPLQDVLAARQELIELDAVTPG-------DERVGRALAGLQSAS
Rv0845|M.tuberculosis_H37Rv AEAIHDGPLQDVLAARQELIELDAVTPG-------DERVGRALAGLQSAS
MMAR_4781|M.marinum_M SESIHDGPLQDVLAARQELIELRAAAPH-------DQRVSTALAGLQSAS
MUL_0350|M.ulcerans_Agy99 SESIHDGPLQDVLAARQELIELRAAAPH-------DQRVSTALAGLQSAS
MAV_0874|M.avium_104 SEFIHDGPLQDILAVRQELVELDAAVPD-------DERIGRALAGLQLAS
TH_4203|M.thermoresistible__bu AADIHDGISQRLVTLSYRLDAAIRAVD---DPATVTEQLSKARELADLTL
Mvan_1357|M.vanbaalenii_PYR-1 AGDIHDGISQRLVTLSYRLDAAAQAATSPGDPDALHEQLDRARELAELTL
MLBr_00774|M.leprae_Br4923 TSDVSHELRTPLTTVRMAADLIYDHSSD------LDPTLRRSTELMVSEL
: : . : : :
Mflv_0729|M.gilvum_PYR-GCK DGLRATVTELHPQVLAQLGLTAGIRELLRQLQSRH-LLEVDAELEEVG--
MSMEG_0106|M.smegmatis_MC2_155 TGLRSTVTALHPQVLAQLGLAPAVRELLRQYENRG-DLVIRANIADVG--
MAB_0061|M.abscessus_ATCC_1997 AELRGAVSSLHPQVLIELGLTAAIRELARQHTARG-IVDVSTAVEEVG--
Mb0868|M.bovis_AF2122/97 ERLRQATFELHPAVLEQVGLGPAVKQLAASTAQRS-GIKISTDIDYPI--
Rv0845|M.tuberculosis_H37Rv ERLRQATFELHPAVLEQVGLGPAVKQLAASTAQRS-GIKISTDIDYPI--
MMAR_4781|M.marinum_M DLLRQATFELHPAVLEQVGLGAAVEQLASFHAQRS-GIRITDDVDYPI--
MUL_0350|M.ulcerans_Agy99 DLLRQATFELHPAVLEQVGLGAAVEQLASFHAQRS-GIRITDDVDYPI--
MAV_0874|M.avium_104 ERLRQATFELHPAVLEQVGLGAAVQQLAEFTAQRS-GIEISTDIDYSG--
TH_4203|M.thermoresistible__bu EETRAAISGLRPPVLDDLGLSGGLASLAQSVGQLDIGVDIEVDLAETR--
Mvan_1357|M.vanbaalenii_PYR-1 QEARAAISGLRPPVLDDLGLSGGLASLARTIPQ----VTIDVDLVDTR--
MLBr_00774|M.leprae_Br4923 DRFETLLNDLLEISRHDAGVAELSVEAVDLRVMVNNALGNVGHLAEEAGI
. * : *: . : :
Mflv_0729|M.gilvum_PYR-GCK -----------EPQ-SQALLYRAARELLTNIVKHADATTVRVRLAR----
MSMEG_0106|M.smegmatis_MC2_155 -----------QPE-AQALLYRAARELLANVHKHARAATVTVRLFR----
MAB_0061|M.abscessus_ATCC_1997 -----------SPP-AQPLVYRAAKELLTNAVKHGRAKNIRVELSS----
Mb0868|M.bovis_AF2122/97 -----------RSG-IDPIVFGVVRELLSNVVRHSGATTASVRLGI----
Rv0845|M.tuberculosis_H37Rv -----------RSG-IDPIVFGVVRELLSNVVRHSGATTASVRLGI----
MMAR_4781|M.marinum_M -----------RDE-IDPIVFGVARELLSNVVQHSRARNASVALGI----
MUL_0350|M.ulcerans_Agy99 -----------RDE-IDPIVFGVARELLSNVVQHSRARNASVALGI----
MAV_0874|M.avium_104 -----------RSA-IDAVVFGVVRELLSNVVQHSRARTAAIMLGR----
TH_4203|M.thermoresistible__bu -----------LPDHIELALYRIAQECLQNVVKHARARTARLTYAVDRGE
Mvan_1357|M.vanbaalenii_PYR-1 -----------LPDHIELALYRIAQECLQNVVKHARASRARLTFALDRGD
MLBr_00774|M.leprae_Br4923 ELLVDMPVDEVIAEVDARRVERILRNLIANAIDHSEHKPVRIRMAA----
: :: : * *. :
Mflv_0729|M.gilvum_PYR-GCK RGDRVVLSVADDGAGFDP-------------AILGTSLG-QGHIGLGSLI
MSMEG_0106|M.smegmatis_MC2_155 QGERTVLTVVDDGVGFDP-------------DIVAQSVA-EGHIGLASLQ
MAB_0061|M.abscessus_ATCC_1997 RRCWLTLVVTDDGRGFDP-------------RDLVLSVA-DGHIGLASLT
Mb0868|M.bovis_AF2122/97 TDEKCVLDVADDGVGVTG-------------DTMARRLG-EGHIGLASHR
Rv0845|M.tuberculosis_H37Rv TDEKCVLDVADDGVGVTG-------------DTMARRLG-EGHIGLASHR
MMAR_4781|M.marinum_M SDGVCVLDVADDGVGVSG-------------ETMARRLG-QGHIGLASHR
MUL_0350|M.ulcerans_Agy99 SDGVCVLDVADDGVGVSG-------------ETMARRLG-QGHIGLASHR
MAV_0874|M.avium_104 TDGTCVLHVVDDGVGFNQ-------------ETVARRLG-EGHIGLASHR
TH_4203|M.thermoresistible__bu HGEVARLEIVDDGVGFDT-------------FEHPLGGDDMSGYGLLSMA
Mvan_1357|M.vanbaalenii_PYR-1 HGDLARLEIADDGIGFDT-------------LEHPLGGDEMGGYGLLSMA
MLBr_00774|M.leprae_Br4923 DEDTVAVTVRDYGIGLRPGEEKLVFSRFWRSDPSRVRRSGGTGLGLAISI
: : * * *. **
Mflv_0729|M.gilvum_PYR-GCK ARFDAMGGSMRITSSDGQGTEVTATSPPEPA-------------------
MSMEG_0106|M.smegmatis_MC2_155 VRIEAMGGAMTISSEEGFGTQVTVIL------------------------
MAB_0061|M.abscessus_ATCC_1997 VPIEAAGGAVTITSAPGAGTRVSVTAPADMAA------------------
Mb0868|M.bovis_AF2122/97 ARVDAAGGVLVFLATP-RGTHVCVELPLKR--------------------
Rv0845|M.tuberculosis_H37Rv ARVDAAGGVLVFLATP-RGTHVCVELPLKR--------------------
MMAR_4781|M.marinum_M ARVDAAGGSLVFLNTP-TGTHVCVRIPLKG--------------------
MUL_0350|M.ulcerans_Agy99 ARVDAAGGSLVFLNTP-TGTHVCVRIPLKG--------------------
MAV_0874|M.avium_104 ARVEAAGGELVFLDVP-VGTHVCVEVPLRN--------------------
TH_4203|M.thermoresistible__bu ERAEIVGGRLNIRSRPGAGTAVIATIPLPPATR-----------------
Mvan_1357|M.vanbaalenii_PYR-1 ERAEIVGGRLNIRSRPGAGTTVTATIPLPANRE-----------------
MLBr_00774|M.leprae_Br4923 EDARLHQGRLEAWGEPGQGACFRLTLPLVRGHKVTTSPLPMKPILQPSPQ
* : *: .
Mflv_0729|M.gilvum_PYR-GCK -----------------------
MSMEG_0106|M.smegmatis_MC2_155 -----------------------
MAB_0061|M.abscessus_ATCC_1997 -----------------------
Mb0868|M.bovis_AF2122/97 -----------------------
Rv0845|M.tuberculosis_H37Rv -----------------------
MMAR_4781|M.marinum_M -----------------------
MUL_0350|M.ulcerans_Agy99 -----------------------
MAV_0874|M.avium_104 -----------------------
TH_4203|M.thermoresistible__bu -----------------------
Mvan_1357|M.vanbaalenii_PYR-1 -----------------------
MLBr_00774|M.leprae_Br4923 ASTAGQQHGTQRQRLREHAERSR