For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTAVTALPTAEKLRTRVRDALAAVGARIALGVPTAHGLPASTPITGDVLFTVPASSAADVDAAVGEAAD AFSTWRTTPAPVRGTLVARLGQLLVEHKSALASLVTIEAGKITSEALGEVQEMIDICEFAVGLSRQLYGR TIASERPGHRLMENWHPLGVVGVITAFNFPVAVWAWNTAVALVCGNTVVWKPSELTPLTAVACQALLERA CADVGAPPAVGRLVLGEREVGEKLVDDPRVSLVSATGSVRMGREVGPRVAARFGRVLLELGGNNAAVVTP AADLDLAVRGIVFSAAGTAGQRCTTLRRLIVHSSIADELVSRIVKANEQLPVGDPFRGGTLVGPLIHTRA YRDMVGALQQAQAEGGEVIGGERVDVGDEGAFYVTPAVVRMPEQSAVVHQETFAPILYVLTYDTLDEAIA LNNAVPQGLSSSIFTLDMREAERFLAADGSDCGIANVNIGTSGAEIGGAFGGEKETGGGRESGSDSWKAY MRRATNTVNYSSDLPLAQGVEFG
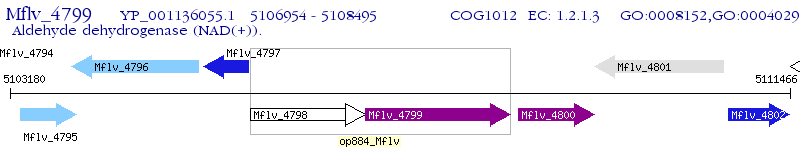
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4799 | - | - | 100% (513) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0357 | - | 1e-58 | 33.88% (484) | methylmalonate-semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0643 | - | 6e-54 | 34.34% (463) | aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3321 | pcd | 0.0 | 76.01% (496) | piperideine-6-carboxilic acid dehydrogenase |
| M. tuberculosis H37Rv | Rv3293 | pcd | 0.0 | 76.21% (496) | piperideine-6-carboxilic acid dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 3e-41 | 30.54% (442) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3649 | - | 0.0 | 79.57% (509) | aldehyde dehydrogenase |
| M. marinum M | MMAR_1240 | pcd | 0.0 | 75.15% (503) | piperideine-6-carboxilic acid dehydrogenase Pcd |
| M. avium 104 | MAV_4265 | - | 0.0 | 77.01% (509) | aldehyde dehydrogenase (NAD) family protein |
| M. smegmatis MC2 155 | MSMEG_1762 | - | 0.0 | 82.75% (510) | piperideine-6-carboxylic acid dehydrogenase |
| M. thermoresistible (build 8) | TH_4137 | - | 2e-57 | 33.70% (454) | PUTATIVE putative [NADP+] succinate-semialdehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_2647 | pcd | 0.0 | 74.75% (503) | piperideine-6-carboxilic acid dehydrogenase Pcd |
| M. vanbaalenii PYR-1 | Mvan_1643 | - | 0.0 | 90.27% (514) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4799|M.gilvum_PYR-GCK MSTAVT----------ALPTAEKLRTRVRDALAAVGARIALGVP---TAH
Mvan_1643|M.vanbaalenii_PYR-1 MTTVVTP---------VLPTADALRTRVRDALAAVGATVPLGAP---GSE
MAB_3649|M.abscessus_ATCC_1997 MTTTLPAPSP-----GVLPSADELRTRAQNALRWIGADVELNTEPPVGDQ
MSMEG_1762|M.smegmatis_MC2_155 MTANITTDKPQGAGAAGLPTADELRQRVRRALDAIGATTALAEPGGASDG
Mb3321|M.bovis_AF2122/97 ---------------------------MLEACQAIGVTAALGEP---GEH
Rv3293|M.tuberculosis_H37Rv ---------------------------MLEACQAIGVTAALGEP---GEH
MMAR_1240|M.marinum_M -----------------MTHSDELRNRVRKAFEAIGVTASLDEP---GGH
MUL_2647|M.ulcerans_Agy99 -----------------MTHSDELRNRVRKAFEAIGVTASLDEP---GGH
MAV_4265|M.avium_104 ---------------MTLPTADELRARAADALRSLGAHVELGAP---DAH
MLBr_02573|M.leprae_Br4923 -------------------------------------------------M
TH_4137|M.thermoresistible__bu --------------------------------MYIDGEWQSAAS----GK
Mflv_4799|M.gilvum_PYR-GCK GLPASTPITGDVLFTVPASSAADVDAAVGEAADAFSTWRTTPAPVRGTLV
Mvan_1643|M.vanbaalenii_PYR-1 GLPASTPITGDVLFSVPQASAADADAAVAAAAEAFSTWRTTPAPVRGALV
MAB_3649|M.abscessus_ATCC_1997 GVTVRTPITGDTLFILTASSKDDVDTAIAEAAQAFSQWRTTPGPIRGALV
MSMEG_1762|M.smegmatis_MC2_155 DLHASTPITGDVLFTLPAHTAEAADAAIAEAAQAFSVWRTTPAPVRGALV
Mb3321|M.bovis_AF2122/97 SLPASTPITGDVLFSIAPTTPEQADHAIAAAAATFTAWRSTPAPVRGALV
Rv3293|M.tuberculosis_H37Rv SLPASTPITGDVLFSIAPTTPEQADHAIAAAAATFTAWRSTPAPVRGALV
MMAR_1240|M.marinum_M GMPSSTPITCKTLFTLTATTAEQTERSIADAQAAFTQWRSTPAPVRGALV
MUL_2647|M.ulcerans_Agy99 GMPSSTPITCETLFTLTATTAEQTERSIADAQAAFTQWRSTPAPVRGALV
MAV_4265|M.avium_104 GLPASTPITGEVLFTVAPTTPDQAEHAITQAAQAFSEWRRTPAPVRGALV
MLBr_02573|M.leprae_Br4923 SIATINPATGETVKTFTPTTQDEVEDAIARAYARFDNYRHTTFTQRAKWA
TH_4137|M.thermoresistible__bu TFASINPANGAVLDSVPDGEREDARRAIDAAAAALPDWRSRTAYERAEVL
. .* . .: .. . :: * : :* . *.
Mflv_4799|M.gilvum_PYR-GCK ARLGQLLVEHKSALASLVTIEAGKITSEALGEVQEMIDICEFAVGLSRQL
Mvan_1643|M.vanbaalenii_PYR-1 ARLGQLLVEHKTALATLVTIEAGKINSEALGEVQEMIDICEFAVGLSRQL
MAB_3649|M.abscessus_ATCC_1997 ARLGELLVEHKKDLAELVTIEAGKIVSEALGEVQEMIDICQFAVGLSRQL
MSMEG_1762|M.smegmatis_MC2_155 ARLGQLLVEHKADLATLVTVEAGKITSEALGEVQEMIDICDFAVGLSRQL
Mb3321|M.bovis_AF2122/97 ARLGELLTAHQQDLATLVTVEVGKITAEARGEVQEMIDVCQFSVGLSRQL
Rv3293|M.tuberculosis_H37Rv ARLGELLTAHQQDLATLVTVEVGKITAEARGEVQEMIDVCQFSVGLSRQL
MMAR_1240|M.marinum_M ARLGELISAHKHDLAELVTIEVGKTGSEALGEVQEMIDICEFAVGLSRQL
MUL_2647|M.ulcerans_Agy99 ARLGELISAHKHDLAELVTIEVGKTGSEALGEVQEMIDICEFAVGLSRQL
MAV_4265|M.avium_104 ARLGELLRAHRDELAALVTLEVGKITSEAAGEVQEMIDICQFAVGLSRQL
MLBr_02573|M.leprae_Br4923 NATADLLEAEANQSAALMTLEMGKTLQSAKDEAMKCAKGFRYYAEKAETL
TH_4137|M.thermoresistible__bu VEAHRIMLERRDDLAELMTREQGKPLRMARTEVGYAADFLLWFAEEAKRV
:: . * *:* * ** * *. . : . :. :
Mflv_4799|M.gilvum_PYR-GCK YGRTIASERPG--HRLMENWHPLGVVGVITAFNFPVAVWAWNTAVALVCG
Mvan_1643|M.vanbaalenii_PYR-1 YGRTIASERPG--HRLMENWHPLGVVGVITAFNFPVAVWAWNTAIALVCG
MAB_3649|M.abscessus_ATCC_1997 YGKTMASERAG--HRLMESWHPLGVVGVISAFNFPVAVWSWNTAIALVCG
MSMEG_1762|M.smegmatis_MC2_155 YGRTIASERPG--HRLMETWHPLGVVGVITAFNFPVAVWSWNTAIALVCG
Mb3321|M.bovis_AF2122/97 YGRTIASERAG--HRLLETWHPLGVVGVITAFNFPVAVWAWNTAVALVCG
Rv3293|M.tuberculosis_H37Rv YGRTIASERAG--HRLLETWHPLGVVGVITAFNFPVAVWAWNTAVALVCG
MMAR_1240|M.marinum_M YGLTIASERPG--HRLMETWHPLGVVGVITAFNFPVAVWAWNTAVALVCG
MUL_2647|M.ulcerans_Agy99 YGLTIASERPG--HRLMETWHPLGVVGVITAFNFPVAVWAWNTAVALVCG
MAV_4265|M.avium_104 YGRTIASERPG--HRLSETWHPLGVVGVITAFNFPVAVWAWNAALALVCG
MLBr_02573|M.leprae_Br4923 LADEPAEAAAVGASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAG
TH_4137|M.thermoresistible__bu YGEVIPSARAD--QRFIVLAQPVGVCAAITPWNYPISMLTRKLAPALAAG
. .. . : :*:** .: .:*:*: * ** .*
Mflv_4799|M.gilvum_PYR-GCK NTVVWKPSELTPLTAVACQALLERACADVGAPPAVGRLVLGEREVGEKLV
Mvan_1643|M.vanbaalenii_PYR-1 DTVVWKPSELTPLTAVACQALLERACADVGAPAAVGRLVLGEREVGEKLV
MAB_3649|M.abscessus_ATCC_1997 DTVVWKPSELTPLTAVACQALLDRAAADVGAPPQVSRLIQGGREVGEQLV
MSMEG_1762|M.smegmatis_MC2_155 DTVVWKPSELTPLTAIACQALIERAAADVGAPAAVSRLVQGGREVGERLV
Mb3321|M.bovis_AF2122/97 DTVVWKPSELTPLTALACQALLSRAAADVGAPAAVGGLLLGGAERGAQLV
Rv3293|M.tuberculosis_H37Rv DTVVWKPSELTPLTALACQALLSRAAADVGAPAAVGGLLLGGAERGAQLV
MMAR_1240|M.marinum_M DTVLWKPSELAPLTALACQALLTRAAADVGAPPSVGAVVLGGAELGEQLV
MUL_2647|M.ulcerans_Agy99 DTVLWKPSELAPLTALACQALLTRAAADVGAPPSVGAVVLGGAELGEQLV
MAV_4265|M.avium_104 DTVVWKPSELTPLTALATQALIARAAADVGAPPGVSAVVLGDRELGELLV
MLBr_02573|M.leprae_Br4923 NVGLLKHASNVPQCALYLADVIARGGFPEDCFQTLLISANG----VEAIL
TH_4137|M.thermoresistible__bu CTSVLKPAEQTPLCAVETFKVFADAGVPAGVVNLVTCSDPEP--VADELL
. : * :. .* *: :: . . : ::
Mflv_4799|M.gilvum_PYR-GCK DDPRVSLVSATGSVRMGREVGPRVAARFGRVLLELGGNNAAVVTPAADLD
Mvan_1643|M.vanbaalenii_PYR-1 DDPRVALVSATGSVRMGREVGPRVAARFGRALLELGGNNAAIVTPAADLD
MAB_3649|M.abscessus_ATCC_1997 DDPRIALVSATGSVRMGQQVGPRVAARFGRSLLELGGNNAAIVTPSADLD
MSMEG_1762|M.smegmatis_MC2_155 DDPRVALVSATGSVRMGREVGPRVAARFGKVLLELGGNNAAIVTPSADLD
Mb3321|M.bovis_AF2122/97 DDPRVALLSATGSVRMGQQVGPRVARRFGRVLLELGGNNAAIVAPSADLE
Rv3293|M.tuberculosis_H37Rv DDPRVALLSATGSVRMGQQVGPRVARRFGRVLLELGGNNAAIVAPSADLE
MMAR_1240|M.marinum_M DDPRIALLSATGSVRMGQQVGPRVARRFGRVLLELGGNNAVIVTPSADLD
MUL_2647|M.ulcerans_Agy99 DDPRIALLSATGSVRMGQQVGPRVARRFGRVLLELGGNNAVIVTPSADLD
MAV_4265|M.avium_104 DDERVALVSATGSVRMGRAVGPRVAARFGRVLLELGGNNAAVVTPSADLD
MLBr_02573|M.leprae_Br4923 RDPRVAAATLTGSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLD
TH_4137|M.thermoresistible__bu GDPRVRKISFTGSTEVGRMIASRAGATMKRVSMELGGHAPFLVFPDADPE
* *: : *** *: :.. .. : :**** . :* * ** :
Mflv_4799|M.gilvum_PYR-GCK LAVRGIVFSAAGTAGQRCTTLRRLIVHSSIADELVSRIVKANEQLPVGDP
Mvan_1643|M.vanbaalenii_PYR-1 LAVRGIVFSAAGTAGQRCTTLRRLIVHSSVADELVSRIVAAYKQLPVGDP
MAB_3649|M.abscessus_ATCC_1997 LAVRGIVFSAAGTAGQRCTTLRRLIVHRSIADGLVERIVHAYGQLPVGSP
MSMEG_1762|M.smegmatis_MC2_155 LAVRAIVFSAAGTAGQRCTTLRRLIVHSSVADEVVRRVVSAYQSLPIGDP
Mb3321|M.bovis_AF2122/97 LAVRCIVFAAAGTAGQRCTSLRRLIVHRSVADDVVARVVGAYRQLAIGDP
Rv3293|M.tuberculosis_H37Rv LAVRGIVFAAAGTAGQRCTSLRRLIVHRSVADDVVARVVGAYRQLAIGDP
MMAR_1240|M.marinum_M LAVRGIVFAAAGTAGQRCTSLRRLIVHSSVADDVVARVVAACRTLPVGDP
MUL_2647|M.ulcerans_Agy99 LAVRGIVFAAAGTAGQRCTSLRRLIVHSSVADDVVARVVAACRTLPVGDP
MAV_4265|M.avium_104 LAVRAVVFAAAGTAGQRCTTLRRLIVHHSVADEVVARVVSALRRLPIGDP
MLBr_02573|M.leprae_Br4923 AAVATAVTGRVQNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDP
TH_4137|M.thermoresistible__bu HAAKGGAAVKFLNTGQACICPNRFIVHRSIAEPFIATLRARIEKLVPGDG
*. . . ** * .*:*.* .: : .: . * *.
Mflv_4799|M.gilvum_PYR-GCK FRGGTLVGPLIHTRAYRDMVGALQQAQAEGGEVIGGERVD--VGDE-GAF
Mvan_1643|M.vanbaalenii_PYR-1 FADRTLVGPLIHTASYRDMVLALQQATAEGGTVLGGERVD--VGDE-GAF
MAB_3649|M.abscessus_ATCC_1997 FDEQTLIGPLINDKAFRDMQDALETARAQGGTVVGGERRE--LGD--GSF
MSMEG_1762|M.smegmatis_MC2_155 SAEGTLVGPLIHARAYRDMVGALEQARADGGTVHGGERHE--LGDEEGAF
Mb3321|M.bovis_AF2122/97 SAPDTLVGPLIHEAAYRDMVAALERARTDGGEVIGGDRRE---VGSPGAY
Rv3293|M.tuberculosis_H37Rv SAPDTLVGPLIHEAAYRDMVAALERARTDGGEVIGGDRRE---VGSPGAY
MMAR_1240|M.marinum_M CDPATLIGPLIHETAYRDMLGALDQARAEGGEVIDPNPRR---DGPAGGY
MUL_2647|M.ulcerans_Agy99 CDPATLIGPLIHETAYRDMLGALDQARAEGGEVIDPNPRR---DGPAGGY
MAV_4265|M.avium_104 FEPGTLVGPLIHETAYRDMVGALESARADGGEVIDGDRRHPFAVEHPSSY
MLBr_02573|M.leprae_Br4923 TDPDTDVGPLATESGRAQVEQQVDAAAAAGAVIRCGGKRPDRP----GWF
TH_4137|M.thermoresistible__bu LAAGTTVGPLIDEVALKKMQRQVDDAVDKGAKLELGGSRLQGPAFDNGHF
* :*** . .: :: * *. : . :
Mflv_4799|M.gilvum_PYR-GCK YVTPAVVRMPEQSAVVHQETFAPILYVLTYDTLDEAIALNNAVPQGLSSS
Mvan_1643|M.vanbaalenii_PYR-1 YVTPAVVRMPAQSAVVHKETFAPILYVMTYETLDEAIALNNAVPQGLSSS
MAB_3649|M.abscessus_ATCC_1997 YVTPAVVRMPEQSDVVHRETFAPILYVLDYDTLDEAIALNNAVPQGLSSA
MSMEG_1762|M.smegmatis_MC2_155 YVAPAVVVMPAQTEIVHNETFAPILYVLTYDDLDEAIALNNAVPQGLSSA
Mb3321|M.bovis_AF2122/97 YVAPAVVRMPSQTAIVATETFAPILYVLTYDDLDEAIALNNAVPQGLSSS
Rv3293|M.tuberculosis_H37Rv YVAPAVVRMPSQTAIVATETFAPILYVLTYDDLDEAIALNNAVPQGLSSS
MMAR_1240|M.marinum_M YVAPAVVRMPAQTAIVATETFAPILYVLTYDDLDDAIALNNAVPQGLSSA
MUL_2647|M.ulcerans_Agy99 YVAPAVVRMPAQTAIVATETFAPILYVLTYDDLDDAIALNNAVPQGLSSA
MAV_4265|M.avium_104 YVAPAVVRMPAQTPIVATETFAPILYVLTYDRLDDAIALNNAVPQGLSSA
MLBr_02573|M.leprae_Br4923 YPPTVITDITKDMALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSN
TH_4137|M.thermoresistible__bu YAPTILTGVTPDMLICSEETFGPIAPVMVFDDEDEAIAMANSTEYGLAGY
* .. :. :. : : *.*.*: * *:** : *:. **..
Mflv_4799|M.gilvum_PYR-GCK IFTLDMREAERFLAADGSDCGIANVN-IGTSGAEIGGAFGGEKETGGGRE
Mvan_1643|M.vanbaalenii_PYR-1 IFTLDMREAERFLAADGSDCGIANVN-IGTSGAEIGGAFGGEKETGGGRE
MAB_3649|M.abscessus_ATCC_1997 IFTLDMREAERFLAADGSDCGIANVN-IGTSGAEIGGAFGGEKETGGGRE
MSMEG_1762|M.smegmatis_MC2_155 IFTTDVREAERFLAADGSDCGIANVN-IGTSGAEIGGAFGGEKQTGGGRE
Mb3321|M.bovis_AF2122/97 IFTTDLREAEHFLDQ--SDCGIANVN-IGTSGAEIGGAFGGEKQTGGGRE
Rv3293|M.tuberculosis_H37Rv IFTTDLREAEHFLDQ--SDCGIANVN-IGTSGAEIGGAFGGEKQTGGGRE
MMAR_1240|M.marinum_M IFTNDLREAERFLAA--SDCGIANVN-IGTSGAEIGGAFGGEKQTGGGRE
MUL_2647|M.ulcerans_Agy99 FFTNDLREAERFLAA--SDCGIANVN-IGTSGAEIGGAFGGEKQTGGGRE
MAV_4265|M.avium_104 IFTTDLREAERFLDE--SDCGIANVN-IGTSGAEIGGAFGGEKQTGGGRE
MLBr_02573|M.leprae_Br4923 AWTQNESEQRHFIDN--IVAGQVFINGMTVSYPELP--FGGIKRSGYGRE
TH_4137|M.thermoresistible__bu IYTRDISRAIRVGEA--LDFGMIGINDINPTSAAVP--FGGIKQSGLGRE
:* : . :. * :* : : . : *** *.:* ***
Mflv_4799|M.gilvum_PYR-GCK SGSDSWKAYMRRATNTVNYSSDLPLAQGVEFG
Mvan_1643|M.vanbaalenii_PYR-1 SGSDSWKAYMRRATNTVNYSTELPLAQGVHFG
MAB_3649|M.abscessus_ATCC_1997 SGSDSWKAYMRRVTNTVNYSTELPLAQGVHFG
MSMEG_1762|M.smegmatis_MC2_155 SGSDAWKAYMRRATNTVNYSSELPLAQGVHFG
Mb3321|M.bovis_AF2122/97 SGSDAWKAYMRRATNTVNYSSELPLAQGVKFG
Rv3293|M.tuberculosis_H37Rv SGSDAWKAYMRRATNTVNYSSELPLAQGVKFG
MMAR_1240|M.marinum_M SGSDAWKAYMRRATNTINYSTELPLAQGVQFG
MUL_2647|M.ulcerans_Agy99 SGSDAWKAYMRRATNTINYSTELPLAQSVQFG
MAV_4265|M.avium_104 SGSDSWKAYMRQSTNTVNYSGALPLAQGVEFG
MLBr_02573|M.leprae_Br4923 LSTHGIREFCNIKTVWIA--------------
TH_4137|M.thermoresistible__bu GARAGIEEYLDTKVLGIAL-------------
. . . : . :