For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTHSDELRNRVRKAFEAIGVTASLDEPGGHGMPSSTPITCETLFTLTATTAEQTERSIADAQAAFTQWRS TPAPVRGALVARLGELISAHKHDLAELVTIEVGKTGSEALGEVQEMIDICEFAVGLSRQLYGLTIASERP GHRLMETWHPLGVVGVITAFNFPVAVWAWNTAVALVCGDTVLWKPSELAPLTALACQALLTRAAADVGAP PSVGAVVLGGAELGEQLVDDPRIALLSATGSVRMGQQVGPRVARRFGRVLLELGGNNAVIVTPSADLDLA VRGIVFAAAGTAGQRCTSLRRLIVHSSVADDVVARVVAACRTLPVGDPCDPATLIGPLIHETAYRDMLGA LDQARAEGGEVIDPNPRRDGPAGGYYVAPAVVRMPAQTAIVATETFAPILYVLTYDDLDDAIALNNAVPQ GLSSAFFTNDLREAERFLAASDCGIANVNIGTSGAEIGGAFGGEKQTGGGRESGSDAWKAYMRRATNTIN YSTELPLAQSVQFG
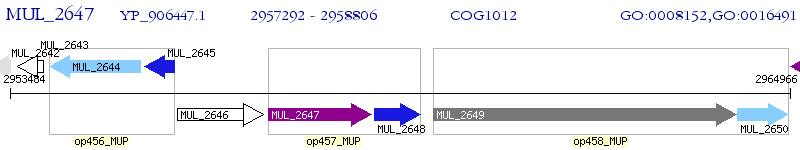
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2647 | pcd | - | 100% (504) | piperideine-6-carboxilic acid dehydrogenase Pcd |
| M. ulcerans Agy99 | MUL_2789 | - | 6e-53 | 33.49% (436) | gamma-aminobutyraldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_0837 | mmsA | 2e-48 | 30.79% (445) | methylmalonate semialdehyde dehydrogenase, MmsA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3321 | pcd | 0.0 | 80.77% (494) | piperideine-6-carboxilic acid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4799 | - | 0.0 | 74.75% (503) | aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv3293 | pcd | 0.0 | 80.97% (494) | piperideine-6-carboxilic acid dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 3e-50 | 31.83% (443) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3649 | - | 0.0 | 74.31% (506) | aldehyde dehydrogenase |
| M. marinum M | MMAR_1240 | pcd | 0.0 | 99.40% (504) | piperideine-6-carboxilic acid dehydrogenase Pcd |
| M. avium 104 | MAV_4265 | - | 0.0 | 77.58% (504) | aldehyde dehydrogenase (NAD) family protein |
| M. smegmatis MC2 155 | MSMEG_1762 | - | 0.0 | 77.12% (507) | piperideine-6-carboxylic acid dehydrogenase |
| M. thermoresistible (build 8) | TH_4531 | - | 1e-61 | 34.38% (448) | PUTATIVE putative betaine aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1643 | - | 0.0 | 74.95% (503) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4799|M.gilvum_PYR-GCK MSTAVT----------ALPTAEKLRTRVRDALAAVGARIALGVP---TAH
Mvan_1643|M.vanbaalenii_PYR-1 MTTVVTP---------VLPTADALRTRVRDALAAVGATVPLGAP---GSE
MAB_3649|M.abscessus_ATCC_1997 MTTTLPAPSP-----GVLPSADELRTRAQNALRWIGADVELNTEPPVGDQ
MSMEG_1762|M.smegmatis_MC2_155 MTANITTDKPQGAGAAGLPTADELRQRVRRALDAIGATTALAEPGGASDG
MUL_2647|M.ulcerans_Agy99 -----------------MTHSDELRNRVRKAFEAIGVTASLDEP---GGH
MMAR_1240|M.marinum_M -----------------MTHSDELRNRVRKAFEAIGVTASLDEP---GGH
Mb3321|M.bovis_AF2122/97 ---------------------------MLEACQAIGVTAALGEP---GEH
Rv3293|M.tuberculosis_H37Rv ---------------------------MLEACQAIGVTAALGEP---GEH
MAV_4265|M.avium_104 ---------------MTLPTADELRARAADALRSLGAHVELGAP---DAH
TH_4531|M.thermoresistible__bu --------------------VTTTENRLVSEIHLAGRRLAEGSAG-----
MLBr_02573|M.leprae_Br4923 -------------------------------------------------M
Mflv_4799|M.gilvum_PYR-GCK GLPASTPITGDVLFTVPASSAADVDAAVGEAADAFSTWRTTPAPVRGTLV
Mvan_1643|M.vanbaalenii_PYR-1 GLPASTPITGDVLFSVPQASAADADAAVAAAAEAFSTWRTTPAPVRGALV
MAB_3649|M.abscessus_ATCC_1997 GVTVRTPITGDTLFILTASSKDDVDTAIAEAAQAFSQWRTTPGPIRGALV
MSMEG_1762|M.smegmatis_MC2_155 DLHASTPITGDVLFTLPAHTAEAADAAIAEAAQAFSVWRTTPAPVRGALV
MUL_2647|M.ulcerans_Agy99 GMPSSTPITCETLFTLTATTAEQTERSIADAQAAFTQWRSTPAPVRGALV
MMAR_1240|M.marinum_M GMPSSTPITCKTLFTLTATTAEQTERSIADAQAAFTQWRSTPAPVRGALV
Mb3321|M.bovis_AF2122/97 SLPASTPITGDVLFSIAPTTPEQADHAIAAAAATFTAWRSTPAPVRGALV
Rv3293|M.tuberculosis_H37Rv SLPASTPITGDVLFSIAPTTPEQADHAIAAAAATFTAWRSTPAPVRGALV
MAV_4265|M.avium_104 GLPASTPITGEVLFTVAPTTPDQAEHAITQAAQAFSEWRRTPAPVRGALV
TH_4531|M.thermoresistible__bu VYEHVNPATGKVQGEVPLAGPAEVDEAVAAAQEAFKEWRKWRPEDRRRLL
MLBr_02573|M.leprae_Br4923 SIATINPATGETVKTFTPTTQDEVEDAIARAYARFDNYRHTTFTQRAKWA
.* * .. .. .: :: * * :* *
Mflv_4799|M.gilvum_PYR-GCK ARLGQLLVEHKSALASLVTIEAGKITSEALG-EVQEMIDICEFAVGLSRQ
Mvan_1643|M.vanbaalenii_PYR-1 ARLGQLLVEHKTALATLVTIEAGKINSEALG-EVQEMIDICEFAVGLSRQ
MAB_3649|M.abscessus_ATCC_1997 ARLGELLVEHKKDLAELVTIEAGKIVSEALG-EVQEMIDICQFAVGLSRQ
MSMEG_1762|M.smegmatis_MC2_155 ARLGQLLVEHKADLATLVTVEAGKITSEALG-EVQEMIDICDFAVGLSRQ
MUL_2647|M.ulcerans_Agy99 ARLGELISAHKHDLAELVTIEVGKTGSEALG-EVQEMIDICEFAVGLSRQ
MMAR_1240|M.marinum_M ARLGELISAHKHDLAELVTIEVGKTGSEALG-EVQEMIDICEFAVGLSRQ
Mb3321|M.bovis_AF2122/97 ARLGELLTAHQQDLATLVTVEVGKITAEARG-EVQEMIDVCQFSVGLSRQ
Rv3293|M.tuberculosis_H37Rv ARLGELLTAHQQDLATLVTVEVGKITAEARG-EVQEMIDVCQFSVGLSRQ
MAV_4265|M.avium_104 ARLGELLRAHRDELAALVTLEVGKITSEAAG-EVQEMIDICQFAVGLSRQ
TH_4531|M.thermoresistible__bu TRIAERIRAHEDELAVLQARENGLPVSQGKAVLVPHGAGWFEAAAGWADK
MLBr_02573|M.leprae_Br4923 NATADLLEAEANQSAALMTLEMGKTLQSAKDEAMKCAKGFRYYAEKAETL
.: : . * * : * * .. : . :
Mflv_4799|M.gilvum_PYR-GCK LYGRTIASERP-GHRLMENWHPLGVVGVITAFNFPVAVWAWNTAVALVCG
Mvan_1643|M.vanbaalenii_PYR-1 LYGRTIASERP-GHRLMENWHPLGVVGVITAFNFPVAVWAWNTAIALVCG
MAB_3649|M.abscessus_ATCC_1997 LYGKTMASERA-GHRLMESWHPLGVVGVISAFNFPVAVWSWNTAIALVCG
MSMEG_1762|M.smegmatis_MC2_155 LYGRTIASERP-GHRLMETWHPLGVVGVITAFNFPVAVWSWNTAIALVCG
MUL_2647|M.ulcerans_Agy99 LYGLTIASERP-GHRLMETWHPLGVVGVITAFNFPVAVWAWNTAVALVCG
MMAR_1240|M.marinum_M LYGLTIASERP-GHRLMETWHPLGVVGVITAFNFPVAVWAWNTAVALVCG
Mb3321|M.bovis_AF2122/97 LYGRTIASERA-GHRLLETWHPLGVVGVITAFNFPVAVWAWNTAVALVCG
Rv3293|M.tuberculosis_H37Rv LYGRTIASERA-GHRLLETWHPLGVVGVITAFNFPVAVWAWNTAVALVCG
MAV_4265|M.avium_104 LYGRTIASERP-GHRLSETWHPLGVVGVITAFNFPVAVWAWNAALALVCG
TH_4531|M.thermoresistible__bu LEGAVVPTTPG-ETVDLTLLEPIGVVAVILTWNAPIGGWGMCVAPPLAAG
MLBr_02573|M.leprae_Br4923 LADEPAEAAAVGASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAG
* . : .*:*** .: .:* *: .* .* .*
Mflv_4799|M.gilvum_PYR-GCK NTVVWKPSELTPLTAVACQALLERACADVGAPPAVGRLVLGEREVGEKLV
Mvan_1643|M.vanbaalenii_PYR-1 DTVVWKPSELTPLTAVACQALLERACADVGAPAAVGRLVLGEREVGEKLV
MAB_3649|M.abscessus_ATCC_1997 DTVVWKPSELTPLTAVACQALLDRAAADVGAPPQVSRLIQGGREVGEQLV
MSMEG_1762|M.smegmatis_MC2_155 DTVVWKPSELTPLTAIACQALIERAAADVGAPAAVSRLVQGGREVGERLV
MUL_2647|M.ulcerans_Agy99 DTVLWKPSELAPLTALACQALLTRAAADVGAPPSVGAVVLGGAELGEQLV
MMAR_1240|M.marinum_M DTVLWKPSELAPLTALACQALLTRAAADVGAPPSVGAVVLGGAELGEQLV
Mb3321|M.bovis_AF2122/97 DTVVWKPSELTPLTALACQALLSRAAADVGAPAAVGGLLLGGAERGAQLV
Rv3293|M.tuberculosis_H37Rv DTVVWKPSELTPLTALACQALLSRAAADVGAPAAVGGLLLGGAERGAQLV
MAV_4265|M.avium_104 DTVVWKPSELTPLTALATQALIARAAADVGAPPGVSAVVLGDRELGELLV
TH_4531|M.thermoresistible__bu CCVILKPSELAPFTS----SFIARICEEEGMPPGVVTVLPGGPDAGEALV
MLBr_02573|M.leprae_Br4923 NVGLLKHASNVPQCALYLADVIARG----GFPEDCFQTLLISANGVEAIL
: * :. .* : .: * * * : : ::
Mflv_4799|M.gilvum_PYR-GCK DDPRVSLVSATGSVRMGREVGPRVAARFGRVLLELGGNNAAVVTPAADLD
Mvan_1643|M.vanbaalenii_PYR-1 DDPRVALVSATGSVRMGREVGPRVAARFGRALLELGGNNAAIVTPAADLD
MAB_3649|M.abscessus_ATCC_1997 DDPRIALVSATGSVRMGQQVGPRVAARFGRSLLELGGNNAAIVTPSADLD
MSMEG_1762|M.smegmatis_MC2_155 DDPRVALVSATGSVRMGREVGPRVAARFGKVLLELGGNNAAIVTPSADLD
MUL_2647|M.ulcerans_Agy99 DDPRIALLSATGSVRMGQQVGPRVARRFGRVLLELGGNNAVIVTPSADLD
MMAR_1240|M.marinum_M DDPRIALLSATGSVRMGQQVGPRVARRFGRVLLELGGNNAVIVTPSADLD
Mb3321|M.bovis_AF2122/97 DDPRVALLSATGSVRMGQQVGPRVARRFGRVLLELGGNNAAIVAPSADLE
Rv3293|M.tuberculosis_H37Rv DDPRVALLSATGSVRMGQQVGPRVARRFGRVLLELGGNNAAIVAPSADLE
MAV_4265|M.avium_104 DDERVALVSATGSVRMGRAVGPRVAARFGRVLLELGGNNAAVVTPSADLD
TH_4531|M.thermoresistible__bu RHPGVAKVSFTGGGPTGRKIAAACGEQLKPVLLELGGKSANIVFPDADLA
MLBr_02573|M.leprae_Br4923 RDPRVAAATLTGSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLD
. :: : **. *: :.. . .: :*****... :* * ***
Mflv_4799|M.gilvum_PYR-GCK LAVRGIVFSAAGTAGQRCTTLRRLIVHSSIADELVSRIVKANEQLPVGDP
Mvan_1643|M.vanbaalenii_PYR-1 LAVRGIVFSAAGTAGQRCTTLRRLIVHSSVADELVSRIVAAYKQLPVGDP
MAB_3649|M.abscessus_ATCC_1997 LAVRGIVFSAAGTAGQRCTTLRRLIVHRSIADGLVERIVHAYGQLPVGSP
MSMEG_1762|M.smegmatis_MC2_155 LAVRAIVFSAAGTAGQRCTTLRRLIVHSSVADEVVRRVVSAYQSLPIGDP
MUL_2647|M.ulcerans_Agy99 LAVRGIVFAAAGTAGQRCTSLRRLIVHSSVADDVVARVVAACRTLPVGDP
MMAR_1240|M.marinum_M LAVRGIVFAAAGTAGQRCTSLRRLIVHSSVADDVVARVVAACRTLPVGDP
Mb3321|M.bovis_AF2122/97 LAVRCIVFAAAGTAGQRCTSLRRLIVHRSVADDVVARVVGAYRQLAIGDP
Rv3293|M.tuberculosis_H37Rv LAVRGIVFAAAGTAGQRCTSLRRLIVHRSVADDVVARVVGAYRQLAIGDP
MAV_4265|M.avium_104 LAVRAVVFAAAGTAGQRCTTLRRLIVHHSVADEVVARVVSALRRLPIGDP
TH_4531|M.thermoresistible__bu KAA-ETAASVIFSAGQACTLPTRILVHESIYDEFTAMVAGALGQIPLGDP
MLBr_02573|M.leprae_Br4923 AAVATAVTGRVQNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDP
*. . . . ** * *::.* .: * .. .. : :*.*
Mflv_4799|M.gilvum_PYR-GCK FRGGTLVGPLIHTRAYRDMVGALQQAQAE-GGEVIGGERVD--VGDE-GA
Mvan_1643|M.vanbaalenii_PYR-1 FADRTLVGPLIHTASYRDMVLALQQATAE-GGTVLGGERVD--VGDE-GA
MAB_3649|M.abscessus_ATCC_1997 FDEQTLIGPLINDKAFRDMQDALETARAQ-GGTVVGGERRE--LGD--GS
MSMEG_1762|M.smegmatis_MC2_155 SAEGTLVGPLIHARAYRDMVGALEQARAD-GGTVHGGERHE--LGDEEGA
MUL_2647|M.ulcerans_Agy99 CDPATLIGPLIHETAYRDMLGALDQARAE-GGEVIDPNPRR---DGPAGG
MMAR_1240|M.marinum_M CDPATLIGPLIHETAYRDMLGALDQARAE-GGEVIDPNPRR---DGPAGG
Mb3321|M.bovis_AF2122/97 SAPDTLVGPLIHEAAYRDMVAALERARTD-GGEVIGGDRRE---VGSPGA
Rv3293|M.tuberculosis_H37Rv SAPDTLVGPLIHEAAYRDMVAALERARTD-GGEVIGGDRRE---VGSPGA
MAV_4265|M.avium_104 FEPGTLVGPLIHETAYRDMVGALESARAD-GGEVIDGDRRHPFAVEHPSS
TH_4531|M.thermoresistible__bu LDPATMMGPVISEAAVERILRTVDEATRDNGAELLTGGSRAG--GELASG
MLBr_02573|M.leprae_Br4923 TDPDTDVGPLATESGRAQVEQQVDAAAAA-GAVIRCGGKRP-----DRPG
* :**: . : :: * *. : .
Mflv_4799|M.gilvum_PYR-GCK FYVTPAVVR-MPEQSAVVHQETFAPILYVLTYDTLDEAIALNNAVPQGLS
Mvan_1643|M.vanbaalenii_PYR-1 FYVTPAVVR-MPAQSAVVHKETFAPILYVMTYETLDEAIALNNAVPQGLS
MAB_3649|M.abscessus_ATCC_1997 FYVTPAVVR-MPEQSDVVHRETFAPILYVLDYDTLDEAIALNNAVPQGLS
MSMEG_1762|M.smegmatis_MC2_155 FYVAPAVVV-MPAQTEIVHNETFAPILYVLTYDDLDEAIALNNAVPQGLS
MUL_2647|M.ulcerans_Agy99 YYVAPAVVR-MPAQTAIVATETFAPILYVLTYDDLDDAIALNNAVPQGLS
MMAR_1240|M.marinum_M YYVAPAVVR-MPAQTAIVATETFAPILYVLTYDDLDDAIALNNAVPQGLS
Mb3321|M.bovis_AF2122/97 YYVAPAVVR-MPSQTAIVATETFAPILYVLTYDDLDEAIALNNAVPQGLS
Rv3293|M.tuberculosis_H37Rv YYVAPAVVR-MPSQTAIVATETFAPILYVLTYDDLDEAIALNNAVPQGLS
MAV_4265|M.avium_104 YYVAPAVVR-MPAQTPIVATETFAPILYVLTYDRLDDAIALNNAVPQGLS
TH_4531|M.thermoresistible__bu YYIEPTLLGNVSPTSEVAQNEVFGPVTAAIRFSDEEEAVAIANGTPYGLA
MLBr_02573|M.leprae_Br4923 WFYPPTVITDITKDMALYTDEVFGPVASVYCATNIDDAIEIANATPFGLG
:: *::: :. : *.*.*: . ::*: : *..* **.
Mflv_4799|M.gilvum_PYR-GCK SSIFTLDMREAERFLAADGSDCGIANVNIGTSGAEIGGAFGGEKETGGGR
Mvan_1643|M.vanbaalenii_PYR-1 SSIFTLDMREAERFLAADGSDCGIANVNIGTSGAEIGGAFGGEKETGGGR
MAB_3649|M.abscessus_ATCC_1997 SAIFTLDMREAERFLAADGSDCGIANVNIGTSGAEIGGAFGGEKETGGGR
MSMEG_1762|M.smegmatis_MC2_155 SAIFTTDVREAERFLAADGSDCGIANVNIGTSGAEIGGAFGGEKQTGGGR
MUL_2647|M.ulcerans_Agy99 SAFFTNDLREAERFLAA--SDCGIANVNIGTSGAEIGGAFGGEKQTGGGR
MMAR_1240|M.marinum_M SAIFTNDLREAERFLAA--SDCGIANVNIGTSGAEIGGAFGGEKQTGGGR
Mb3321|M.bovis_AF2122/97 SSIFTTDLREAEHFLDQ--SDCGIANVNIGTSGAEIGGAFGGEKQTGGGR
Rv3293|M.tuberculosis_H37Rv SSIFTTDLREAEHFLDQ--SDCGIANVNIGTSGAEIGGAFGGEKQTGGGR
MAV_4265|M.avium_104 SAIFTTDLREAERFLDE--SDCGIANVNIGTSGAEIGGAFGGEKQTGGGR
TH_4531|M.thermoresistible__bu GYVHTRDVSRVLRLASA--LEAGTVAFNGAGAPAGPGAPFGGYKESGYGR
MLBr_02573|M.leprae_Br4923 SNAWTQNESEQRHFIDN--IVAGQVFING-MTVSYPELPFGGIKRSGYGR
. * : . :: .* . .* : : .*** *.:* **
Mflv_4799|M.gilvum_PYR-GCK ESGSDSWKAYMRRATNTVNYSSDLPLAQGVEFG
Mvan_1643|M.vanbaalenii_PYR-1 ESGSDSWKAYMRRATNTVNYSTELPLAQGVHFG
MAB_3649|M.abscessus_ATCC_1997 ESGSDSWKAYMRRVTNTVNYSTELPLAQGVHFG
MSMEG_1762|M.smegmatis_MC2_155 ESGSDAWKAYMRRATNTVNYSSELPLAQGVHFG
MUL_2647|M.ulcerans_Agy99 ESGSDAWKAYMRRATNTINYSTELPLAQSVQFG
MMAR_1240|M.marinum_M ESGSDAWKAYMRRATNTINYSTELPLAQGVQFG
Mb3321|M.bovis_AF2122/97 ESGSDAWKAYMRRATNTVNYSSELPLAQGVKFG
Rv3293|M.tuberculosis_H37Rv ESGSDAWKAYMRRATNTVNYSSELPLAQGVKFG
MAV_4265|M.avium_104 ESGSDSWKAYMRQSTNTVNYSGALPLAQGVEFG
TH_4531|M.thermoresistible__bu EGGKLGVLEFVQHKTVSVALS------------
MLBr_02573|M.leprae_Br4923 ELSTHGIREFCNIKTVWIA--------------
* .. . : . * :