For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRTQTGRYTSSPHRHAPPPQNKCMAETISHWMNNAPFAGASEATAPVTNPATGAVTGQVALASVADAQAV IDAAAAAFPAWRDTSLAKRTQVLFRFRELLNDRKGELAEIITAEHGKVVSDALGEVSRGQEVVEFACGIP HLLKGSYTENASTKVDVHSLRQPLGPVAIISPFNFPAMVPMWFFPVAIAAGNTVVLKPSEKDPSASLWLA ELWKEAGLPDGVFNVLQGDKTAVDELLTNPRIKSVSFVGSTPIAQYVYATGTAAGKRVQALGGAKNHAVV LPDADLDLAADAMVNAGFGSAGERCMAISACVAVGPIADDLVAKIRERTVGLKIGDGTKDSDMGPLVTRA HRDKVASYIDAGESAGAKVVVDGRAVKADGGQEGFWLGPTLLDNVTPEMSVYTDEIFGPVLSVVRVDTYD QALDLINSNPYGNGTAIFTNDGGAARRFQNEVEVGMVGINVPIPVPMAYYSFGGWKASLFGDSHAHGMDG VHFFTRQKAITTRWLDPSHGGINLGFPENS
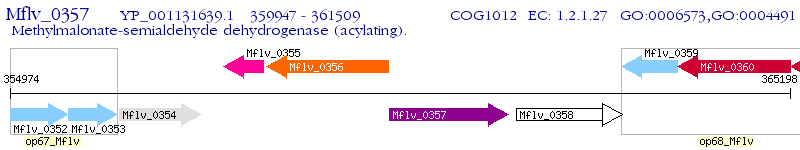
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0357 | - | - | 100% (520) | methylmalonate-semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_5010 | - | e-146 | 53.47% (490) | methylmalonate-semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4860 | - | 2e-63 | 35.23% (457) | gamma-aminobutyraldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0775c | mmsA | 1e-142 | 51.76% (510) | methylmalonate-semialdehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0753c | mmsA | 1e-143 | 51.96% (510) | methylmalonate-semialdehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 2e-51 | 30.80% (448) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4346c | - | 0.0 | 80.04% (496) | methylmalonate-semialdehyde dehydrogenase (MmsA) |
| M. marinum M | MMAR_1079 | mmsA | 1e-144 | 53.47% (490) | methylmalonate semialdehyde dehydrogenase, MmsA |
| M. avium 104 | MAV_4417 | mmsA | 1e-148 | 54.29% (490) | methylmalonate-semialdehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2449 | mmsA | 0.0 | 87.30% (496) | methylmalonate-semialdehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_2909 | - | 1e-144 | 51.29% (503) | PUTATIVE methylmalonate-semialdehyde dehydrogenase [acylating) |
| M. ulcerans Agy99 | MUL_0837 | mmsA | 1e-144 | 53.27% (490) | methylmalonate semialdehyde dehydrogenase, MmsA |
| M. vanbaalenii PYR-1 | Mvan_1824 | - | 0.0 | 89.80% (500) | methylmalonate-semialdehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0357|M.gilvum_PYR-GCK MRTQTGRYTSSPHRHAPPPQNKCMAETISHWMNNAPFAGASEATAPVTNP
Mvan_1824|M.vanbaalenii_PYR-1 -----------------------MTATISHWVNGEVFTGTSSASAPVTNP
MSMEG_2449|M.smegmatis_MC2_155 -----------------------MSTVIQHWRDGKVFAGSSTATAPVTNP
MAB_4346c|M.abscessus_ATCC_199 -----------------------MTQSITHWMNGKTYPGVGGRLAPVTNP
Mb0775c|M.bovis_AF2122/97 -----------------------MTTQISHFIDGQRTAGQSTRSADVFDP
Rv0753c|M.tuberculosis_H37Rv -----------------------MTTQISHFIDGQRTAGQSTRSADVFDP
MAV_4417|M.avium_104 -----------------------MTTQIPHFIDGQRTAGQSTRTAEVFDP
MMAR_1079|M.marinum_M -----------------------MTTQIPHFIDGRRTPGQSTRTADVLDP
MUL_0837|M.ulcerans_Agy99 -----------------------MTTQIPHFIDGRRTPGQSTRTADVLDP
TH_2909|M.thermoresistible__bu -----------------------MTRQIHHFINGQHTAGQSTRTADVMNP
MLBr_02573|M.leprae_Br4923 ------------------------------------------MSIATINP
. :*
Mflv_0357|M.gilvum_PYR-GCK ATGAVTGQVALASVADAQAVIDAAAAAFPAWRDTSLAKRTQVLFRFRELL
Mvan_1824|M.vanbaalenii_PYR-1 ATGTVTGQVALASVDDARAVIDAAAAAFPAWRDASLAKRTSVLFRFRELL
MSMEG_2449|M.smegmatis_MC2_155 ATGAVTGEVALASVEDARAVIDSAAAAFPAWRDTSLAKRTQVLFAFRELL
MAB_4346c|M.abscessus_ATCC_199 ATGAVTGEVVLGSVADARAVIDSATVAFPAWRDMSLARRSAILFNFRELL
Mb0775c|M.bovis_AF2122/97 NTGQIQAKVPMAGKSDIDAAVASAVEAQKGWAAWNPQRRARVLMRFIELV
Rv0753c|M.tuberculosis_H37Rv NTGQIQAKVPMAGKSDIDAAVASAVEAQKGWAAWNPQRRARVLMRFIELV
MAV_4417|M.avium_104 STGAVQAQVPMAGRADIDAAVASAIEAQKGWAATNPQRRARVLMRFIELV
MMAR_1079|M.marinum_M NTGQVQATVPMAAQADIDAAVAGAAEAQIGWAAWNPQRRARVLMRFIELV
MUL_0837|M.ulcerans_Agy99 NTGQVQATVPMAAQADIDAAVAGAAEAQVGWAAWNPQRRARVLMRFIELV
TH_2909|M.thermoresistible__bu STGKVQAKVVMGTAADIDAAVAGAAEAQKEWAAWNPQRRARVLMKFIDLV
MLBr_02573|M.leprae_Br4923 ATGETVKTFTPTTQDEVEDAIARAYARFDNYRHTTFTQRAKWANATADLL
** . : .: * : . :*: :*:
Mflv_0357|M.gilvum_PYR-GCK NDRKGELAEIITAEHGKVVSDALGEVSRGQEVVEFACGIPHLLKGSYTEN
Mvan_1824|M.vanbaalenii_PYR-1 NERKGELAEIITAEHGKVVSDALGEVSRGQEVVEFACGIPHLLKGGFTEN
MSMEG_2449|M.smegmatis_MC2_155 NARKEEVAAIITAEHGKVLSDALGEVTRGLEVVEFACGIPHLLKGGFTEN
MAB_4346c|M.abscessus_ATCC_199 NARKEELAAIITSEHGKVLSDALGEVGRGQEVVEFACGIPHLLKGGFTEN
Mb0775c|M.bovis_AF2122/97 NDTIDELAELLSREHGKTLADARGDVQRGIEVIEFCLGIPHLLKGEYTEG
Rv0753c|M.tuberculosis_H37Rv NDTIDELAELLSREHGKTLADARGDVQRGIEVIEFCLGIPHLLKGEYTEG
MAV_4417|M.avium_104 NQHNDELAELLSREHGKTLADAKGDIQRGIEVIEFCVGIPHLLKGEYSEG
MMAR_1079|M.marinum_M NQNTDELAELLSREHGKTVPDARGDIQRGIEVIEFCLGIPHLLKGEYTEG
MUL_0837|M.ulcerans_Agy99 NQNTDELAELLSREHGKTVPDARGDIQRGIEVIEFCLGIPHLLKGEYTEG
TH_2909|M.thermoresistible__bu NQNVDELAELLSLEHGKTLADAKGDIQRGIEVIEFSIGIPHLQKGEYTEG
MLBr_02573|M.leprae_Br4923 EAEANQSAALMTLEMGKTLQSAKDEAMKCAKGFRYYAEKAETLLADEPAE
: : * ::: * **.: .* .: : : ..: .. . .
Mflv_0357|M.gilvum_PYR-GCK ASTK--VDVHSLRQPLGPVAIISPFNFPAMVPMWFFPVAIAAGNTVVLKP
Mvan_1824|M.vanbaalenii_PYR-1 ASTK--VDVYSIRQPLGPVAVISPFNFPAMVPMWFFPIAIATGNTVVLKP
MSMEG_2449|M.smegmatis_MC2_155 ASTK--VDVYSISQPLGPVGIISPFNFPAMVPMWFFPIAIAAGNTVVVKP
MAB_4346c|M.abscessus_ATCC_199 ASTN--VDVYSVRQPVGPVAIISPFNFPAMVPLWFIPVALAAGNTVVLKP
Mb0775c|M.bovis_AF2122/97 AGPG--IDVYSLRQPLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKP
Rv0753c|M.tuberculosis_H37Rv AGPG--IDVYSLRQPLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKP
MAV_4417|M.avium_104 AGPG--IDVYSLRQPLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKP
MMAR_1079|M.marinum_M AGPG--IDVYSLRQPLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKP
MUL_0837|M.ulcerans_Agy99 AGPG--IDVYSLRQPLGVVAGITPFNFPAMIPLWKAGPALACGNAFVLKP
TH_2909|M.thermoresistible__bu AGPG--IDVYSMRQPLGVVAGITPFNFPAMIPLWKAGPALACGNAFILKP
MLBr_02573|M.leprae_Br4923 AAAVGASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKH
*.. .. : **:* * : *:*** .: *: **. ::*
Mflv_0357|M.gilvum_PYR-GCK SEKDPSASLWLAELWKEAGLPDGVFNVLQGDKTAVDELLTNPRIKSVSFV
Mvan_1824|M.vanbaalenii_PYR-1 SEKDPSASLWLAALWKEAGLPDGVFNVLQGDKTAVDELLANPKIKSVSFV
MSMEG_2449|M.smegmatis_MC2_155 SEKDPSASLWMAELWKEAGLPDGVFNVLQGDKTAVDELLTNPKIKGVSFV
MAB_4346c|M.abscessus_ATCC_199 SEKDPSASLWLARLLAEAGVPAGVFNVLQGDKTVVDELLTHPAVKAVSFV
Mb0775c|M.bovis_AF2122/97 SERDPSVPVRLAELFIEAGLPAGVFQVVHGDKEAVDAILHHPDIKAVGFV
Rv0753c|M.tuberculosis_H37Rv SERDPSVPVRLAELFIEAGLPAGVFQVVHGDKEAVDAILHHPDIKAVGFV
MAV_4417|M.avium_104 SERDPSVPVRLAELFVEAGLPPGVFQVVHGDKEAVDAILNHPDIQAVGFV
MMAR_1079|M.marinum_M SERDPSVPLRLAELFVEAGLPPGVFQVVHGDKEAVDAILNHPEIKAVGFV
MUL_0837|M.ulcerans_Agy99 SERDPSVPLRLAELFVEAGLPPGVFQVVHGDKEAVDAILNHPEIKAVGFV
TH_2909|M.thermoresistible__bu SERDPSVPVRLAELFLQAGLPSGLFQVVHGDKEAVDAILEHPTIQAVGFV
MLBr_02573|M.leprae_Br4923 ASNVPQCALYLADVIARGGFPEDCFQTLLISANGVEAILRDPRVAAATLT
:.. *. .: :* : ..*.* . *:.: . *: :* .* : .. :.
Mflv_0357|M.gilvum_PYR-GCK GSTPIAQYVYATGTAAGKRVQALGGAKNHAVVLPDADLDLAADAMVNAGF
Mvan_1824|M.vanbaalenii_PYR-1 GSTPIAQYVYATGTAAGKRVQALGGAKNHAVILPDADLDLAADAMVNAGF
MSMEG_2449|M.smegmatis_MC2_155 GSTPIAKYVYATGTAAGKRVQALGGAKNHAVVLPDADLDLAADAMVNAGF
MAB_4346c|M.abscessus_ATCC_199 GSTPIAQYVYATASAAGKRVQALGGAKNHAVVLPDADLDLAADALVNAGF
Mb0775c|M.bovis_AF2122/97 GSSDIAQYIYAGAAATGKRAQCFGGAKNHMIVMPDADLDQAVDALIGAGY
Rv0753c|M.tuberculosis_H37Rv GSSDIAQYIYAGAAATGKRAQCFGGAKNHMIVMPDADLDQAVDALIGAGY
MAV_4417|M.avium_104 GSSDIAQYIYAGAAATGKRSQCFGGAKNHMIVMPDADLDQAVDALIGAGY
MMAR_1079|M.marinum_M GSSDIAQYIYSGAAATGKRSQCFGGAKNHMIVMPDADLDQAVDALIGAGY
MUL_0837|M.ulcerans_Agy99 GSSDIAQYIYSGAAATGKRSQCFGGAKNHMIVMPDADLDQAVDALIGAGY
TH_2909|M.thermoresistible__bu GSSDIAQYIYAGATAHGKRAQCFGGAKNHMIVMPDADLDQAVDALIGAGY
MLBr_02573|M.leprae_Br4923 GSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRV
** .: : : . * *..: :::*.**** *. : : .
Mflv_0357|M.gilvum_PYR-GCK GSAGERCMAISACVAVG-PIADDLVAKIRERTVGLKIGDGTK-DSDMGPL
Mvan_1824|M.vanbaalenii_PYR-1 GSAGERCMAISACVAVG-PIADDLVAKIRERTVGLKIGDGTK-ESDMGPL
MSMEG_2449|M.smegmatis_MC2_155 GSAGERCMAISACVAVG-PVADELVAKIAERAATIKTGDGTK-DSDMGPL
MAB_4346c|M.abscessus_ATCC_199 GSAGERCMAISVAVAVG-SIGDELVARIAQRARGIKTGDGTG-DADMGPL
Mb0775c|M.bovis_AF2122/97 GSAGERCMAISVAVPVGDQTAERLRARLIERINNLRVGHSLDPKADYGPL
Rv0753c|M.tuberculosis_H37Rv GSAGERCMAISVAVPVGDQTAERLRARLIERINNLRVGHSLDPKADYGPL
MAV_4417|M.avium_104 GSAGERCMAISVAVPVGDKTADRLRARLVERINNLRVGHSLDPKADYGPL
MMAR_1079|M.marinum_M GSAGERCMAISVAVPVGEQTADRLRAKLVERINNLRVGHSLDPKADYGPL
MUL_0837|M.ulcerans_Agy99 GSTGERCMAISVAVPVGEQTADRLRAKLVERINNLRVGHSLDPKADYGPL
TH_2909|M.thermoresistible__bu GSAGERCMAISVAVPVGDEIADRLRARLIERVANLRVGHSLDPKADYGPL
MLBr_02573|M.leprae_Br4923 QNNGQSCIAAKRFIAHA-DIYDEFVEKFVARMETLRVGDPTDPDTDVGPL
. *: *:* . :. . : : :: * :: *. .:* ***
Mflv_0357|M.gilvum_PYR-GCK VTRAHRDKVASYIDAGESAGAKVVVDGRAVKAD--------GGQE----G
Mvan_1824|M.vanbaalenii_PYR-1 VTQAHRDKVASYIDAGEADGAKVVVDGRSVGSE--------GAPERDPAG
MSMEG_2449|M.smegmatis_MC2_155 VTKAHRDKVASYIDAGEAAGAKVVLDGRTVIAN--------GEED----G
MAB_4346c|M.abscessus_ATCC_199 VTKAHRDRVASYIEAGRAAGAAIVVDGREVEAN--------GASD----G
Mb0775c|M.bovis_AF2122/97 VTGAALARVRDYIGQGVAAGAELVVDGRDRASDDLTFGLPEGDANLEG-G
Rv0753c|M.tuberculosis_H37Rv VTGAALARVRDYIGQGVAAGAELVVDGRDRASDDLTFGLPEGDANLEG-G
MAV_4417|M.avium_104 VTGAAVARVRDYIDQGVAAGAEIVVDGRERRSDDLTF----GDANLEG-G
MMAR_1079|M.marinum_M VTEAALTRVRDYIGQGVASGAELVVDGRERASDDLAF----GDANLEG-G
MUL_0837|M.ulcerans_Agy99 VTEAALTRVRDYIGQGVASGAELVVDGRERASDDLAF----GDANLEG-G
TH_2909|M.thermoresistible__bu VTEAALQRVRDYIGQGVEAGAELLVDGRERGSDDLQF----GDDSLEG-G
MLBr_02573|M.leprae_Br4923 ATESGRAQVEQQVDAAAAAGAVIRCGGKRPDRP----------------G
.* : :* . : . ** : .*: *
Mflv_0357|M.gilvum_PYR-GCK FWLGPTLLDNVTPEMSVYTDEIFGPVLSVVRVDTYDQALDLINSNPYGNG
Mvan_1824|M.vanbaalenii_PYR-1 FWLGPTLLDNVTPEMSVYTDEIFGPVLSVLRVESYDQALELINTNPYGNG
MSMEG_2449|M.smegmatis_MC2_155 FWLGPTLLDNVTPEMSVYTDEIFGPVLSVVRVETYDEALELINNNPYGNG
MAB_4346c|M.abscessus_ATCC_199 FWLGPTLIDHVGPDMSVYTDEIFGPILSVVRVDTYDEALELVNNSPYGNG
Mb0775c|M.bovis_AF2122/97 FFIGPTLFDHVAAHMSIYTDEIFGPVLCMVRARDYEEALRLPSEHEYGNG
Rv0753c|M.tuberculosis_H37Rv FFIGPTLFDHVAAHMSIYTDEIFGPVLCMVRARDYEEALRLPSEHEYGNG
MAV_4417|M.avium_104 FFIGPTLFDHVTPDMSIYTDEIFGPVLCIVRAQNYEEALRLPSEHEYGNG
MMAR_1079|M.marinum_M YFIGPTLFDHVTTDMSIYTDEIFGPVLCMVRAHDYEEALRLPSEHEYGNG
MUL_0837|M.ulcerans_Agy99 YFIGPTLFDHVTTDMSIYTDEIFGPVLCMVRAHDYEEALRLPSEHEYGNG
TH_2909|M.thermoresistible__bu YFIGPTLFDRVTTDMSIYRDEIFGPVLCMVRAKNYEEALALPSEHEYGNG
MLBr_02573|M.leprae_Br4923 WFYPPTVITDITKDMALYTDEVFGPVASVYCATNIDDAIEIANATPFGLG
:: **:: : .*::* **:***: .: . ::*: : . :* *
Mflv_0357|M.gilvum_PYR-GCK TAIFTNDGGAARRFQNEVEVGMVGINVPIPVPMAYYSFGGWKASLFGDSH
Mvan_1824|M.vanbaalenii_PYR-1 TAIFTNDGGAARRFQNEVEVGMVGINVPIPVPMAYFSFGGWKASLFGDSH
MSMEG_2449|M.smegmatis_MC2_155 TAIFTNDGGAARRFQNEVEVGMVGINVPIPVPMAYYSFGGWKASLFGDTH
MAB_4346c|M.abscessus_ATCC_199 TAIFTNDGGAARRFQNEVEVGMVGINVPIPVPMAYYSFGGWKSSLFGDTH
Mb0775c|M.bovis_AF2122/97 VAIFTRDGDAARDFVSRGQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLN
Rv0753c|M.tuberculosis_H37Rv VAIFTRDGDAARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLN
MAV_4417|M.avium_104 VAVFTRDGDAARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLN
MMAR_1079|M.marinum_M VAVFTRDGDTARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLN
MUL_0837|M.ulcerans_Agy99 VAVFTRDGDTARDFVSRVQVGMVGVNVPIPVPVAYHTFGGWKRSGFGDLN
TH_2909|M.thermoresistible__bu VAIFTRDGDTARDFVSRVQCGMVGVNVPIPVPVAYHTFGGWKRSGFGDLN
MLBr_02573|M.leprae_Br4923 SNAWTQNESEQRHFIDNIVAGQVFING-MTVSYPELPFGGIKRSGYG--R
:*.: . * * .. * * :* :.*. . .*** * * :* .
Mflv_0357|M.gilvum_PYR-GCK AHGMDGVHFFTRQKAITTRWLDPSHGGINLGFPENS
Mvan_1824|M.vanbaalenii_PYR-1 AHGMDGVQFFTRQKAVTQRWLDPSHGGINLGFPQNG
MSMEG_2449|M.smegmatis_MC2_155 AHGIQGVHFFTRTKAVTSRWLDPSHGGINLGFPQNK
MAB_4346c|M.abscessus_ATCC_199 AHGMEGVNFYTRGKAITSRWLDPSHGGINLGFPENK
Mb0775c|M.bovis_AF2122/97 QHGPAAIQFYTKVKTVTSRWPSGIKDGAEFVIPTMS
Rv0753c|M.tuberculosis_H37Rv QHGPAAIQFYTKVKTVTSRWPSGIKDGAEFVIPTMS
MAV_4417|M.avium_104 QHGPASIQFYTKVKTVTSRWPSGIKDGAEFVIPTMD
MMAR_1079|M.marinum_M QHGPTSIQFYTKVKTVTSRWPSGIKDGAEFVIPTMK
MUL_0837|M.ulcerans_Agy99 QHGPTSIQFYTKVKTVTSRWPSGIKDGAEFVIPTMK
TH_2909|M.thermoresistible__bu QHGPHSILFYTKTKTVTQRWPSGIKDGAEFSIPTMK
MLBr_02573|M.leprae_Br4923 ELSTHGIREFCNIKTVWIA-----------------
. .: : . *::