For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNLLALAFTAGMLAPVNPCGFALLPAWITGTIATGGTDAVLVRLARALRTGAVLTIGFTGTLTLAGIAIS AGARTLVTAAPWLGITIGLTLAILGGFMLTGRTIGLRMPARTRHRPDSPTAGGVLAAGIGYALASLSCTF GVLLAVIAQAQATSGWGGLLAVFTAYTAGAATILMLVSVGTAIAGTALTRHLGILARHGTRVTAVVLIAT GAYLAWYWLPAATGHTASGGNLLTGWSATATGWLQDHALPASVIAAFAVVVSAVAAYWTTHRRPITGKQR RR
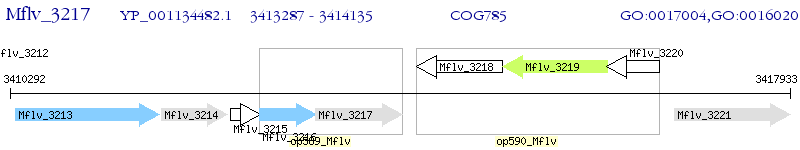
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3217 | - | - | 100% (282) | cytochrome c biogenesis protein, transmembrane region |
| M. gilvum PYR-GCK | Mflv_0051 | - | 3e-10 | 31.00% (229) | cytochrome c biogenesis protein, transmembrane region |
| M. gilvum PYR-GCK | Mflv_2825 | - | 3e-08 | 29.33% (225) | cytochrome c biogenesis protein, transmembrane region |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2902c | - | 8e-29 | 30.22% (278) | integral membrane protein |
| M. tuberculosis H37Rv | Rv2877c | - | 8e-29 | 30.22% (278) | integral membrane protein |
| M. leprae Br4923 | MLBr_02411 | - | 2e-07 | 27.43% (226) | putative cytochrome C-type biogenesis protein |
| M. abscessus ATCC 19977 | MAB_4275c | - | 2e-23 | 32.57% (218) | hypothetical protein MAB_4275c |
| M. marinum M | MMAR_1832 | - | 2e-25 | 27.62% (286) | integral membrane protein |
| M. avium 104 | MAV_3728 | - | 8e-28 | 35.32% (218) | cytochrome C biogenesis protein transmembrane region |
| M. smegmatis MC2 155 | MSMEG_3541 | - | 1e-30 | 32.71% (266) | cytochrome C biogenesis protein transmembrane region |
| M. thermoresistible (build 8) | TH_3805 | - | 7e-16 | 32.14% (140) | PUTATIVE putative NADH-quinone oxidoreductase, L subunit |
| M. ulcerans Agy99 | MUL_2080 | - | 2e-25 | 27.62% (286) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_0867 | - | 1e-09 | 30.40% (227) | cytochrome c biogenesis protein, transmembrane region |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2902c|M.bovis_AF2122/97 -MNEALIGLAFAAGLVAALNPCGFAMLPAYLLLVVHGQDS--AGRTG-PL
Rv2877c|M.tuberculosis_H37Rv -MNEALIGLAFAAGLVAALNPCGFAMLPAYLLLVVYGQDS--AGRTG-PL
MMAR_1832|M.marinum_M -MNSALVALAFAAGSVAALNPCGFAMLPAYLLLVVRGQHGGESGEVGSPI
MUL_2080|M.ulcerans_Agy99 -MNSALVALAFAAGSVAALNPCGFAMLPAYLLLVVRGQHGGESGEAASPI
MAV_3728|M.avium_104 -MDRGLVGLAVAAGMVAALNPCGFAMLPAYLLLVVRG-----AGARAPGV
TH_3805|M.thermoresistible__bu --------------------------------------------------
MSMEG_3541|M.smegmatis_MC2_155 -MEQDLLGLAFGAGLVAALNPCGFALLPGYLALVVRGE------NAATGA
MAB_4275c|M.abscessus_ATCC_199 MIDTAALTFALGAGLVAALNPCGFAFLPGYLGLVIAGSDG-----TASRM
MLBr_02411|M.leprae_Br4923 --MTGFTEIAAAGPLLVALGVCMLAGLVSFVSPCVVP-------LVPGYL
Mvan_0867|M.vanbaalenii_PYR-1 MTLDQIDQLTATGPLLLATGLAALAGLVSFASPCVIP-------LVPGYL
Mflv_3217|M.gilvum_PYR-GCK ---MNLLALAFTAGMLAPVNPCGFALLPAWITGTIATG------GTDAVL
Mb2902c|M.bovis_AF2122/97 SAVGRAAAATVGMALGFLTVFGIFGALTISAATAVQRYLPYATVLIGLAL
Rv2877c|M.tuberculosis_H37Rv SAVGRAAAATVGMALGFLTVFGIFGALTISAATAVQRYLPYATVLIGLAL
MMAR_1832|M.marinum_M GAVGRAIAATIGMALGFLTVFGAFGALTISAASTVQRYLPYLTVLIGVLL
MUL_2080|M.ulcerans_Agy99 GAVGRAIAATIGMALGFLTVFGAFGALTISAASTVQRYLPYLTVSIGVLL
MAV_3728|M.avium_104 AAAGRALAASAGMALGFLTVFGLFGALTVSAAGAVQRWLPYATVVVGVLL
TH_3805|M.thermoresistible__bu ----------------------------------VQRYLPVATVVVGAGL
MSMEG_3541|M.smegmatis_MC2_155 RALGRAALATVAMAAGFVAVFGTFTALTVATASTVQRYLPYVTVVIGLIL
MAB_4275c|M.abscessus_ATCC_199 TAIARAATATVAMAGGFLTVFGVFGLVVSPVVASAGRYMPFATVVIGVVL
MLBr_02411|M.leprae_Br4923 SYLAAVVGVSHEETQPGAGVIKTPPAARWRVAGSAVLFVAGFTTVFVLDT
Mvan_0867|M.vanbaalenii_PYR-1 SYLAAVVGVDQSAAGSAAPGAVAVKGARLRVAGAAGLFVAGFTVVFLLGT
Mflv_3217|M.gilvum_PYR-GCK VRLARALRTGAVLTIGFTGTLTLAGIAISAGARTLVTAAPWLGITIGLTL
. .
Mb2902c|M.bovis_AF2122/97 IALGGWLLLGRGLTALTPRSLG--VRWAPTVRLGSMYGYGISYAVASLSC
Rv2877c|M.tuberculosis_H37Rv IALGGWLLLGRGLTALTPRSLG--VRWAPTVRLGSMYGYGISYAVASLSC
MMAR_1832|M.marinum_M FGLGVWLLSGRELAALTPSGFG--PRWAPTARLGSMYGYGVSYAVASLSC
MUL_2080|M.ulcerans_Agy99 FGLGVWLLSGRELAALTPSGFG--PRWAPTARLGSMYGYGVSYAVASLSC
MAV_3728|M.avium_104 VALGGWLLAGRSLSALTPRPVG--PLWAPTARLGSMYGYGISYAIASLSC
TH_3805|M.thermoresistible__bu VALGFWLLIGRELTGLTSVTRR--VERAPTARIGSMFGYGVSYAVASLSC
MSMEG_3541|M.smegmatis_MC2_155 VCLGIWLLAGRRLGVLMPNSLADNPRWAPTARMGSMLGYGVGYALASLSC
MAB_4275c|M.abscessus_ATCC_199 VALSIWLLCGRELTLVLPRVSG----GAPTASLLSMFGYGLTYAVASLSC
MLBr_02411|M.leprae_Br4923 VAVLGMTTVLTTHQVLLQRVGG-----VLTIVMGLVFVGLLPALQRQVQF
Mvan_0867|M.vanbaalenii_PYR-1 VAVLGMTTALITNQLLLQRLGG-----VVTILMGLVFIGLVPVLQRDTRF
Mflv_3217|M.gilvum_PYR-GCK AILGGFMLTGRTIGLRMPARTR---HRPDSPTAGGVLAAGIGYALASLSC
: : : : .
Mb2902c|M.bovis_AF2122/97 TIGPFLAVTGAGLRGGSVVGSVAIYLAYVAGLTLVVGVLAVAAATASSAL
Rv2877c|M.tuberculosis_H37Rv TIGPFLAVTGAGLRGGSVVGSVAIYLAYVAGLTLVVGVLAVAAATASSAL
MMAR_1832|M.marinum_M TIGPFIAVTGAGLRGGSILGAVAIYLAYIAGLTMVVGALAIAAATASSAL
MUL_2080|M.ulcerans_Agy99 TIGPFIAVTGAGLRGGSILGAVAIYLAYIAGLTMVVGALAIAAATASSAL
MAV_3728|M.avium_104 TVGPFLAVTAAGLRSGSLLTGAAVYLGYVAGLTLVVGVLAVAAATAGAAT
TH_3805|M.thermoresistible__bu TVAPFLAVTGAGLRGQSLWSTLSIYLAYAAGFTLIVGVLAMAAALTHSAL
MSMEG_3541|M.smegmatis_MC2_155 TIGPFLAVTGAGLRSGTPLRGVGVFASYAAGFTLVVGVLAVATALARTVV
MAB_4275c|M.abscessus_ATCC_199 TVGPFLAVISTTFKQGSIVSGVLAFIAYGAGMAVTVGVAALAVALLGNSV
MLBr_02411|M.leprae_Br4923 SLRQLTTVAGAPVLGTVFALGWTPCLG-----PTLSGVITVAAATDGVNV
Mvan_0867|M.vanbaalenii_PYR-1 TPRQISTLGGAPLLGAVFALGWTPCLG-----PTLTGVIAVASATEGSNV
Mflv_3217|M.gilvum_PYR-GCK TFGVLLAVIAQAQATSGWGGLLAVFTAYTAGAATILMLVSVGTAIAGTAL
: : :: . . . ::. *
Mb2902c|M.bovis_AF2122/97 ADRLRRILPFVNRISG-ALLVVVGLYVGYYGLYELRLIAGVGANPQDAVI
Rv2877c|M.tuberculosis_H37Rv ADRLRRILPFVNRISG-ALLVVVGLYVGYYGLYELRLIAGVGANPQDAVI
MMAR_1832|M.marinum_M ADRLRRVLPLVNRISG-GLLVVVGLYVGYYGYYEVRLTSGLLVDPQDRVI
MUL_2080|M.ulcerans_Agy99 ADRLRRVLPLVNRISG-GLLAVVGLYVGYYGYYEVRLTSGLLVDPQDRVI
MAV_3728|M.avium_104 ADRLRRMVPFVDRISG-AVLVLVGLYVAYYGGYEVRLFG-AGGDPRDAVI
TH_3805|M.thermoresistible__bu AVRLRRIGRYVNRISG-VLLVAVGGYVAYYGLYELRLFW-GGANPQDPVI
MSMEG_3541|M.smegmatis_MC2_155 IDRMRRILPYANRISG-ALLILVGAYVGYYGLYEVRLFS-GSGDPHDPVI
MAB_4275c|M.abscessus_ATCC_199 QATMRKVLPYVGRIAG-VIVLLTGLYVAYYGYYEIRLNF-GDGSADDPVI
MLBr_02411|M.leprae_Br4923 TRGILLVIAYCMGLGIPFVLLASGSAQAVAGLRWLRQYG--------RAI
Mvan_0867|M.vanbaalenii_PYR-1 ARGVVLVIAYCLGLGIPFVLLAFGSARAVAGLGWLRRHT--------RTI
Mflv_3217|M.gilvum_PYR-GCK TRHLGILARHGTRVTA-VVLIATGAYLAWYWLPAATGHT-------ASGG
: : : :: * .
Mb2902c|M.bovis_AF2122/97 AAAGRLQGALAGWVNQHGAWPWAVLLVVLVVGAFAGTWFR--RVRR----
Rv2877c|M.tuberculosis_H37Rv AAAGRLQGALAGWVNQHGAWPWAVLLVVLVVGAFAGTWFR--RVRR----
MMAR_1832|M.marinum_M GAAGRIQGELAGWVHQHGIWPWAVALTLLVAGAFSVSWYR--RARR----
MUL_2080|M.ulcerans_Agy99 GAAGRIQGELAGWVHQHGIWPWVVVLTLLVAGAFSVSWYR--RARR----
MAV_3728|M.avium_104 GAAGRLQGALAGWVHRHGLWPWALALLALAAVRFGRARRRGRRADRLPDP
TH_3805|M.thermoresistible__bu EAAARAQGAIAGWVHRHGGGPWLAATAVLMLVVLATGFVA--RIRRSARR
MSMEG_3541|M.smegmatis_MC2_155 AAAGKIQGTLAGWVHQHGALPWAIGLLVLIG--LGLLWRVRIFARRSPAR
MAB_4275c|M.abscessus_ATCC_199 NAAGTVQAWLVGVVDATGVWPLTGGIALIVAVAAASSYAV--RKGR----
MLBr_02411|M.leprae_Br4923 QVFGGMLLIAIGAALIAGVWDDFVSWLRDAVVSDMRVSI-----------
Mvan_0867|M.vanbaalenii_PYR-1 QVLGGVLMILVGAALVTGLWNDFVSWIRDAFVSDVRLPI-----------
Mflv_3217|M.gilvum_PYR-GCK NLLTGWSATATGWLQDHALPASVIAAFAVVVSAVAAYWTTHRRPITGKQR
* .
Mb2902c|M.bovis_AF2122/97 --------------------------------------------------
Rv2877c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1832|M.marinum_M --------------------------------------------------
MUL_2080|M.ulcerans_Agy99 --------------------------------------------------
MAV_3728|M.avium_104 AAAAAPQVADQHHEDQRRESDQKPVLDVADVVAAQHGQQQPEDDQADDVP
TH_3805|M.thermoresistible__bu --------------------------------------------------
MSMEG_3541|M.smegmatis_MC2_155 RATADDAP------------------------------------------
MAB_4275c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3217|M.gilvum_PYR-GCK RR------------------------------------------------
Mb2902c|M.bovis_AF2122/97 --------------------------------------------------
Rv2877c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1832|M.marinum_M --------------------------------------------------
MUL_2080|M.ulcerans_Agy99 --------------------------------------------------
MAV_3728|M.avium_104 GPVVDPAPSPRRRRHLDGVDAGEVLDLGTGHGESPSDRIGLSNPDSGTTA
TH_3805|M.thermoresistible__bu --------------------------------------------------
MSMEG_3541|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4275c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3217|M.gilvum_PYR-GCK --------------------------------------------------
Mb2902c|M.bovis_AF2122/97 -------------------
Rv2877c|M.tuberculosis_H37Rv -------------------
MMAR_1832|M.marinum_M -------------------
MUL_2080|M.ulcerans_Agy99 -------------------
MAV_3728|M.avium_104 GRARGARPHHGPRRWRAAQ
TH_3805|M.thermoresistible__bu -------------------
MSMEG_3541|M.smegmatis_MC2_155 -------------------
MAB_4275c|M.abscessus_ATCC_199 -------------------
MLBr_02411|M.leprae_Br4923 -------------------
Mvan_0867|M.vanbaalenii_PYR-1 -------------------
Mflv_3217|M.gilvum_PYR-GCK -------------------