For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DRGLVGLAVAAGMVAALNPCGFAMLPAYLLLVVRGAGARAPGVAAAGRALAASAGMALGFLTVFGLFGAL TVSAAGAVQRWLPYATVVVGVLLVALGGWLLAGRSLSALTPRPVGPLWAPTARLGSMYGYGISYAIASLS CTVGPFLAVTAAGLRSGSLLTGAAVYLGYVAGLTLVVGVLAVAAATAGAATADRLRRMVPFVDRISGAVL VLVGLYVAYYGGYEVRLFGAGGDPRDAVIGAAGRLQGALAGWVHRHGLWPWALALLALAAVRFGRARRRG RRADRLPDPAAAAAPQVADQHHEDQRRESDQKPVLDVADVVAAQHGQQQPEDDQADDVPGPVVDPAPSPR RRRHLDGVDAGEVLDLGTGHGESPSDRIGLSNPDSGTTAGRARGARPHHGPRRWRAAQ*
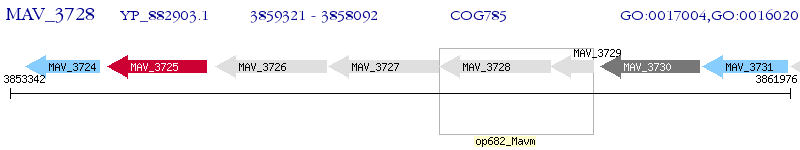
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3728 | - | - | 100% (409) | cytochrome C biogenesis protein transmembrane region |
| M. avium 104 | MAV_4618 | - | 2e-06 | 26.64% (229) | cytochrome C biogenesis protein transmembrane region |
| M. avium 104 | MAV_4839 | - | 4e-06 | 28.45% (232) | hypothetical protein MAV_4839 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2902c | - | 1e-111 | 71.53% (288) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_3217 | - | 1e-27 | 35.32% (218) | cytochrome c biogenesis protein, transmembrane region |
| M. tuberculosis H37Rv | Rv2877c | - | 1e-111 | 71.53% (288) | integral membrane protein |
| M. leprae Br4923 | MLBr_02411 | - | 6e-06 | 26.41% (231) | putative cytochrome C-type biogenesis protein |
| M. abscessus ATCC 19977 | MAB_3244 | - | 2e-71 | 55.42% (240) | mercuric transporter MerT |
| M. marinum M | MMAR_1832 | - | 1e-107 | 67.93% (290) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_3541 | - | 3e-95 | 60.96% (292) | cytochrome C biogenesis protein transmembrane region |
| M. thermoresistible (build 8) | TH_3805 | - | 3e-62 | 60.95% (210) | PUTATIVE putative NADH-quinone oxidoreductase, L subunit |
| M. ulcerans Agy99 | MUL_2080 | - | 1e-106 | 67.24% (290) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_0867 | - | 1e-09 | 27.39% (230) | cytochrome c biogenesis protein, transmembrane region |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2902c|M.bovis_AF2122/97 MNEALIGLAFAAGLVAALNPCGFAMLPAYLLLVVHGQDS--AGRTG-PLS
Rv2877c|M.tuberculosis_H37Rv MNEALIGLAFAAGLVAALNPCGFAMLPAYLLLVVYGQDS--AGRTG-PLS
MMAR_1832|M.marinum_M MNSALVALAFAAGSVAALNPCGFAMLPAYLLLVVRGQHGGESGEVGSPIG
MUL_2080|M.ulcerans_Agy99 MNSALVALAFAAGSVAALNPCGFAMLPAYLLLVVRGQHGGESGEAASPIG
MAV_3728|M.avium_104 MDRGLVGLAVAAGMVAALNPCGFAMLPAYLLLVVRG-----AGARAPGVA
TH_3805|M.thermoresistible__bu --------------------------------------------------
MAB_3244|M.abscessus_ATCC_1997 --------------------------------MVRG-----PGAEIGKSQ
MSMEG_3541|M.smegmatis_MC2_155 MEQDLLGLAFGAGLVAALNPCGFALLPGYLALVVRG-----ENAATG-AR
MLBr_02411|M.leprae_Br4923 -----------------------------------------------MTG
Mvan_0867|M.vanbaalenii_PYR-1 ---------------------------------------------MTLDQ
Mflv_3217|M.gilvum_PYR-GCK --MNLLALAFTAGMLAPVNPCGFALLPAWITGTIAT------GGTDAVLV
Mb2902c|M.bovis_AF2122/97 AVGRAAAATVGMALGFLTVFGIFGALTISAATAVQRYLPYATVLIGLALI
Rv2877c|M.tuberculosis_H37Rv AVGRAAAATVGMALGFLTVFGIFGALTISAATAVQRYLPYATVLIGLALI
MMAR_1832|M.marinum_M AVGRAIAATIGMALGFLTVFGAFGALTISAASTVQRYLPYLTVLIGVLLF
MUL_2080|M.ulcerans_Agy99 AVGRAIAATIGMALGFLTVFGAFGALTISAASTVQRYLPYLTVSIGVLLF
MAV_3728|M.avium_104 AAGRALAASAGMALGFLTVFGLFGALTVSAAGAVQRWLPYATVVVGVLLV
TH_3805|M.thermoresistible__bu ---------------------------------VQRYLPVATVVVGAGLV
MAB_3244|M.abscessus_ATCC_1997 ALARAVIATVVMAGGFVAVFTVFGLLTVSVASVIQRYLPFVTVVFGIGLV
MSMEG_3541|M.smegmatis_MC2_155 ALGRAALATVAMAAGFVAVFGTFTALTVATASTVQRYLPYVTVVIGLILV
MLBr_02411|M.leprae_Br4923 FTEIAAAGPLLVALGVCMLAGLVSFVSPCVVPLVPGYLSYLAAVVGVSHE
Mvan_0867|M.vanbaalenii_PYR-1 IDQLTATGPLLLATGLAALAGLVSFASPCVIPLVPGYLSYLAAVVGVDQS
Mflv_3217|M.gilvum_PYR-GCK RLARALRTGAVLTIGFTGTLTLAGIAISAGARTLVTAAPWLGITIGLTLA
: . .*
Mb2902c|M.bovis_AF2122/97 ALGGWLLLGRGLTALTPRSLGVR--WAPTVRLGSMYGYGISYAVASLSCT
Rv2877c|M.tuberculosis_H37Rv ALGGWLLLGRGLTALTPRSLGVR--WAPTVRLGSMYGYGISYAVASLSCT
MMAR_1832|M.marinum_M GLGVWLLSGRELAALTPSGFGPR--WAPTARLGSMYGYGVSYAVASLSCT
MUL_2080|M.ulcerans_Agy99 GLGVWLLSGRELAALTPSGFGPR--WAPTARLGSMYGYGVSYAVASLSCT
MAV_3728|M.avium_104 ALGGWLLAGRSLSALTPRPVGPL--WAPTARLGSMYGYGISYAIASLSCT
TH_3805|M.thermoresistible__bu ALGFWLLIGRELTGLTSVTRRVE--RAPTARIGSMFGYGVSYAVASLSCT
MAB_3244|M.abscessus_ATCC_1997 ILGLWLLAGRDIIALMPKMLDAN---APTTRLGSMFGYGVGYAVASLSCT
MSMEG_3541|M.smegmatis_MC2_155 CLGIWLLAGRRLGVLMPNSLADNPRWAPTARMGSMLGYGVGYALASLSCT
MLBr_02411|M.leprae_Br4923 ETQPGAGVIKTPPAARWRVAGSAVLFVAGFTTVFVLDTVAVLGMTTVLTT
Mvan_0867|M.vanbaalenii_PYR-1 AAGSAAPGAVAVKGARLRVAGAAGLFVAGFTVVFLLGTVAVLGMTTALIT
Mflv_3217|M.gilvum_PYR-GCK ILGGFMLTGRTIGLRMPARTRHR---PDSPTAGGVLAAGIGYALASLSCT
: .::: *
Mb2902c|M.bovis_AF2122/97 IGPFLAVTGAGLRGGSVVGSVAIYLAYVAGLTLVVGVLAVAAATASSALA
Rv2877c|M.tuberculosis_H37Rv IGPFLAVTGAGLRGGSVVGSVAIYLAYVAGLTLVVGVLAVAAATASSALA
MMAR_1832|M.marinum_M IGPFIAVTGAGLRGGSILGAVAIYLAYIAGLTMVVGALAIAAATASSALA
MUL_2080|M.ulcerans_Agy99 IGPFIAVTGAGLRGGSILGAVAIYLAYIAGLTMVVGALAIAAATASSALA
MAV_3728|M.avium_104 VGPFLAVTAAGLRSGSLLTGAAVYLGYVAGLTLVVGVLAVAAATAGAATA
TH_3805|M.thermoresistible__bu VAPFLAVTGAGLRGQSLWSTLSIYLAYAAGFTLIVGVLAMAAALTHSALA
MAB_3244|M.abscessus_ATCC_1997 IGPFLAVTSTTFESGSLFDGVMVYLAYASGITLVVGTLAVSTALASTMLL
MSMEG_3541|M.smegmatis_MC2_155 IGPFLAVTGAGLRSGTPLRGVGVFASYAAGFTLVVGVLAVATALARTVVI
MLBr_02411|M.leprae_Br4923 HQVLLQRVGGVLTIVMGLVFVGLLPALQRQVQFSLRQLTTVAGAPVLGTV
Mvan_0867|M.vanbaalenii_PYR-1 NQLLLQRLGGVVTILMGLVFIGLVPVLQRDTRFTPRQISTLGGAPLLGAV
Mflv_3217|M.gilvum_PYR-GCK FGVLLAVIAQAQATSGWGGLLAVFTAYTAGAATILMLVSVGTAIAGTALT
:: . : :: . .
Mb2902c|M.bovis_AF2122/97 DRLRRILPFVNRISGALLVVVGLYVGYYGLYELRLIAGVGANPQDAVIAA
Rv2877c|M.tuberculosis_H37Rv DRLRRILPFVNRISGALLVVVGLYVGYYGLYELRLIAGVGANPQDAVIAA
MMAR_1832|M.marinum_M DRLRRVLPLVNRISGGLLVVVGLYVGYYGYYEVRLTSGLLVDPQDRVIGA
MUL_2080|M.ulcerans_Agy99 DRLRRVLPLVNRISGGLLAVVGLYVGYYGYYEVRLTSGLLVDPQDRVIGA
MAV_3728|M.avium_104 DRLRRMVPFVDRISGAVLVLVGLYVAYYGGYEVRLFG-AGGDPRDAVIGA
TH_3805|M.thermoresistible__bu VRLRRIGRYVNRISGVLLVAVGGYVAYYGLYELRLFW-GGANPQDPVIEA
MAB_3244|M.abscessus_ATCC_1997 KAMRRALPYLNRVSGAILIVVGAYVGYYGSYEVRLFH-ANGNPDDPIINA
MSMEG_3541|M.smegmatis_MC2_155 DRMRRILPYANRISGALLILVGAYVGYYGLYEVRLFS-GSGDPHDPVIAA
MLBr_02411|M.leprae_Br4923 FALGWTPCLGPTLSGVITVAAATDGVNVTRGILLVIAYCMGLGIPFVLLA
Mvan_0867|M.vanbaalenii_PYR-1 FALGWTPCLGPTLTGVIAVASATEGSNVARGVVLVIAYCLGLGIPFVLLA
Mflv_3217|M.gilvum_PYR-GCK RHLGILARHGTRVTAVVLIATGAYLAWYWLPAATGHTASGGN-------L
: ::. : .
Mb2902c|M.bovis_AF2122/97 AGRLQGALAG-WVNQHGAWPWAVLLVVLVVGAFAGTWFR--RVRR-----
Rv2877c|M.tuberculosis_H37Rv AGRLQGALAG-WVNQHGAWPWAVLLVVLVVGAFAGTWFR--RVRR-----
MMAR_1832|M.marinum_M AGRIQGELAG-WVHQHGIWPWAVALTLLVAGAFSVSWYR--RARR-----
MUL_2080|M.ulcerans_Agy99 AGRIQGELAG-WVHQHGIWPWVVVLTLLVAGAFSVSWYR--RARR-----
MAV_3728|M.avium_104 AGRLQGALAG-WVHRHGLWPWALALLALAAVRFGRARRRGRRADRLPDPA
TH_3805|M.thermoresistible__bu AARAQGAIAG-WVHRHGGGPWLAATAVLMLVVLATGFVA--RIRRSARR-
MAB_3244|M.abscessus_ATCC_1997 AGKIQRTISG-WVYLHGAWPWLFILG-LVAVGAAIWWRTNVSEPPSN---
MSMEG_3541|M.smegmatis_MC2_155 AGKIQGTLAG-WVHQHGALPWAIGL--LVLIGLGLLWRVRIFARRSPARR
MLBr_02411|M.leprae_Br4923 SGSAQAVAGLRWLRQYGRAIQVFGGMLLIAIGAALIAGVWDDFVSWLRDA
Mvan_0867|M.vanbaalenii_PYR-1 FGSARAVAGLGWLRRHTRTIQVLGGVLMILVGAALVTGLWNDFVSWIRDA
Mflv_3217|M.gilvum_PYR-GCK LTGWSATATG-WLQDHALPASVIAAFAVVVSAVAAYWTTHRRPITGKQRR
*: : : .
Mb2902c|M.bovis_AF2122/97 --------------------------------------------------
Rv2877c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1832|M.marinum_M --------------------------------------------------
MUL_2080|M.ulcerans_Agy99 --------------------------------------------------
MAV_3728|M.avium_104 AAAAPQVADQHHEDQRRESDQKPVLDVADVVAAQHGQQQPEDDQADDVPG
TH_3805|M.thermoresistible__bu --------------------------------------------------
MAB_3244|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3541|M.smegmatis_MC2_155 ATADDAP-------------------------------------------
MLBr_02411|M.leprae_Br4923 VVSDMRVSI-----------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 FVSDVRLPI-----------------------------------------
Mflv_3217|M.gilvum_PYR-GCK R-------------------------------------------------
Mb2902c|M.bovis_AF2122/97 --------------------------------------------------
Rv2877c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1832|M.marinum_M --------------------------------------------------
MUL_2080|M.ulcerans_Agy99 --------------------------------------------------
MAV_3728|M.avium_104 PVVDPAPSPRRRRHLDGVDAGEVLDLGTGHGESPSDRIGLSNPDSGTTAG
TH_3805|M.thermoresistible__bu --------------------------------------------------
MAB_3244|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3541|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3217|M.gilvum_PYR-GCK --------------------------------------------------
Mb2902c|M.bovis_AF2122/97 ------------------
Rv2877c|M.tuberculosis_H37Rv ------------------
MMAR_1832|M.marinum_M ------------------
MUL_2080|M.ulcerans_Agy99 ------------------
MAV_3728|M.avium_104 RARGARPHHGPRRWRAAQ
TH_3805|M.thermoresistible__bu ------------------
MAB_3244|M.abscessus_ATCC_1997 ------------------
MSMEG_3541|M.smegmatis_MC2_155 ------------------
MLBr_02411|M.leprae_Br4923 ------------------
Mvan_0867|M.vanbaalenii_PYR-1 ------------------
Mflv_3217|M.gilvum_PYR-GCK ------------------