For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IDTAALTFALGAGLVAALNPCGFAFLPGYLGLVIAGSDGTASRMTAIARAATATVAMAGGFLTVFGVFGL VVSPVVASAGRYMPFATVVIGVVLVALSIWLLCGRELTLVLPRVSGGAPTASLLSMFGYGLTYAVASLSC TVGPFLAVISTTFKQGSIVSGVLAFIAYGAGMAVTVGVAALAVALLGNSVQATMRKVLPYVGRIAGVIVL LTGLYVAYYGYYEIRLNFGDGSADDPVINAAGTVQAWLVGVVDATGVWPLTGGIALIVAVAAASSYAVRK GR*
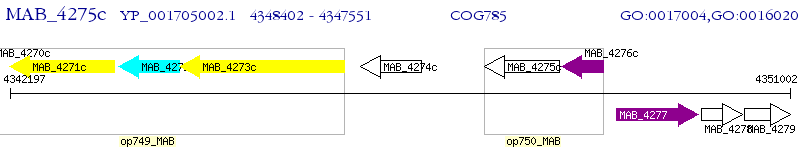
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4275c | - | - | 100% (283) | hypothetical protein MAB_4275c |
| M. abscessus ATCC 19977 | MAB_3244 | - | 2e-62 | 47.72% (241) | mercuric transporter MerT |
| M. abscessus ATCC 19977 | MAB_4286 | - | 3e-07 | 27.67% (206) | putative integral membrane cytochrome c biogenesis protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2902c | - | 7e-70 | 48.43% (287) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_3217 | - | 2e-23 | 32.57% (218) | cytochrome c biogenesis protein, transmembrane region |
| M. tuberculosis H37Rv | Rv2877c | - | 7e-70 | 48.43% (287) | integral membrane protein |
| M. leprae Br4923 | MLBr_02411 | - | 1e-05 | 26.70% (191) | putative cytochrome C-type biogenesis protein |
| M. marinum M | MMAR_5058 | - | 1e-104 | 70.22% (272) | hypothetical protein MMAR_5058 |
| M. avium 104 | MAV_3728 | - | 5e-70 | 47.89% (284) | cytochrome C biogenesis protein transmembrane region |
| M. smegmatis MC2 155 | MSMEG_3541 | - | 4e-68 | 48.54% (274) | cytochrome C biogenesis protein transmembrane region |
| M. thermoresistible (build 8) | TH_3805 | - | 3e-43 | 46.27% (201) | PUTATIVE putative NADH-quinone oxidoreductase, L subunit |
| M. ulcerans Agy99 | MUL_2080 | - | 1e-66 | 47.24% (290) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_0867 | - | 6e-07 | 28.51% (242) | cytochrome c biogenesis protein, transmembrane region |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4275c|M.abscessus_ATCC_199 MIDTAALTFALGAGLVAALNPCGFAFLPGYLGLVIAGS-----DGTASRM
MMAR_5058|M.marinum_M MINTAALSFALGAGLVAALNPCGFAFLPGYLGLVIAGS-----QGTS-RP
Mb2902c|M.bovis_AF2122/97 -MNEALIGLAFAAGLVAALNPCGFAMLPAYLLLVVHGQ---DSAGRTGPL
Rv2877c|M.tuberculosis_H37Rv -MNEALIGLAFAAGLVAALNPCGFAMLPAYLLLVVYGQ---DSAGRTGPL
MUL_2080|M.ulcerans_Agy99 -MNSALVALAFAAGSVAALNPCGFAMLPAYLLLVVRGQHGGESGEAASPI
MAV_3728|M.avium_104 -MDRGLVGLAVAAGMVAALNPCGFAMLPAYLLLVVRGA-----GARAPGV
TH_3805|M.thermoresistible__bu --------------------------------------------------
MSMEG_3541|M.smegmatis_MC2_155 -MEQDLLGLAFGAGLVAALNPCGFALLPGYLALVVRGE------NAATGA
MLBr_02411|M.leprae_Br4923 --MTGFTEIAAAGPLLVALGVCMLAGLVSFVSPCVVPL-------VPGYL
Mvan_0867|M.vanbaalenii_PYR-1 MTLDQIDQLTATGPLLLATGLAALAGLVSFASPCVIPL-------VPGYL
Mflv_3217|M.gilvum_PYR-GCK ---MNLLALAFTAGMLAPVNPCGFALLPAWITGTIATG------GTDAVL
MAB_4275c|M.abscessus_ATCC_199 TAIARAATATVAMAGGFLTVFGVFGLVVSPVVASAGRYMPFATVVIGVVL
MMAR_5058|M.marinum_M RAVLRAGAATAGMSLGFLTVFGVFGLLISPVIASAQRYLPFATVVIGVLL
Mb2902c|M.bovis_AF2122/97 SAVGRAAAATVGMALGFLTVFGIFGALTISAATAVQRYLPYATVLIGLAL
Rv2877c|M.tuberculosis_H37Rv SAVGRAAAATVGMALGFLTVFGIFGALTISAATAVQRYLPYATVLIGLAL
MUL_2080|M.ulcerans_Agy99 GAVGRAIAATIGMALGFLTVFGAFGALTISAASTVQRYLPYLTVSIGVLL
MAV_3728|M.avium_104 AAAGRALAASAGMALGFLTVFGLFGALTVSAAGAVQRWLPYATVVVGVLL
TH_3805|M.thermoresistible__bu ----------------------------------VQRYLPVATVVVGAGL
MSMEG_3541|M.smegmatis_MC2_155 RALGRAALATVAMAAGFVAVFGTFTALTVATASTVQRYLPYVTVVIGLIL
MLBr_02411|M.leprae_Br4923 SYLAAVVGVSHEETQPGAGVIKTPPAARWRVAGSAVLFVAGFTTVFVLDT
Mvan_0867|M.vanbaalenii_PYR-1 SYLAAVVGVDQSAAGSAAPGAVAVKGARLRVAGAAGLFVAGFTVVFLLGT
Mflv_3217|M.gilvum_PYR-GCK VRLARALRTGAVLTIGFTGTLTLAGIAISAGARTLVTAAPWLGITIGLTL
. .
MAB_4275c|M.abscessus_ATCC_199 VALSIWLLCGRELTLVLPRVSG----GAPTASLLSMFGYGLTYAVASLSC
MMAR_5058|M.marinum_M VGLAIWLLAGKELSIILPKSMG----GTPTTRLGSMYGYGVGYAVASLSC
Mb2902c|M.bovis_AF2122/97 IALGGWLLLGRGLTALTPRSLG--VRWAPTVRLGSMYGYGISYAVASLSC
Rv2877c|M.tuberculosis_H37Rv IALGGWLLLGRGLTALTPRSLG--VRWAPTVRLGSMYGYGISYAVASLSC
MUL_2080|M.ulcerans_Agy99 FGLGVWLLSGRELAALTPSGFG--PRWAPTARLGSMYGYGVSYAVASLSC
MAV_3728|M.avium_104 VALGGWLLAGRSLSALTPRPVG--PLWAPTARLGSMYGYGISYAIASLSC
TH_3805|M.thermoresistible__bu VALGFWLLIGRELTGLTSVTRR--VERAPTARIGSMFGYGVSYAVASLSC
MSMEG_3541|M.smegmatis_MC2_155 VCLGIWLLAGRRLGVLMPNSLADNPRWAPTARMGSMLGYGVGYALASLSC
MLBr_02411|M.leprae_Br4923 VAVLGMTTVLTTHQVLLQRVGG-----VLTIVMGLVFVGLLPALQRQVQF
Mvan_0867|M.vanbaalenii_PYR-1 VAVLGMTTALITNQLLLQRLGG-----VVTILMGLVFIGLVPVLQRDTRF
Mflv_3217|M.gilvum_PYR-GCK AILGGFMLTGRTIGLRMPARTR---HRPDSPTAGGVLAAGIGYALASLSC
: : : : .
MAB_4275c|M.abscessus_ATCC_199 TVGPFLAVISTTFKQGSIVSGVLAFIAYGAGMAVTVGVAALAVALLGNSV
MMAR_5058|M.marinum_M TVAPFLAVISTTFKQGSVLSGVLAFVAYAVGMTLTVGVAALTVALAGSSA
Mb2902c|M.bovis_AF2122/97 TIGPFLAVTGAGLRGGSVVGSVAIYLAYVAGLTLVVGVLAVAAATASSAL
Rv2877c|M.tuberculosis_H37Rv TIGPFLAVTGAGLRGGSVVGSVAIYLAYVAGLTLVVGVLAVAAATASSAL
MUL_2080|M.ulcerans_Agy99 TIGPFIAVTGAGLRGGSILGAVAIYLAYIAGLTMVVGALAIAAATASSAL
MAV_3728|M.avium_104 TVGPFLAVTAAGLRSGSLLTGAAVYLGYVAGLTLVVGVLAVAAATAGAAT
TH_3805|M.thermoresistible__bu TVAPFLAVTGAGLRGQSLWSTLSIYLAYAAGFTLIVGVLAMAAALTHSAL
MSMEG_3541|M.smegmatis_MC2_155 TIGPFLAVTGAGLRSGTPLRGVGVFASYAAGFTLVVGVLAVATALARTVV
MLBr_02411|M.leprae_Br4923 SLRQLTTVAGAPVLGTVFALGWTPCLG-----PTLSGVITVAAATDGVNV
Mvan_0867|M.vanbaalenii_PYR-1 TPRQISTLGGAPLLGAVFALGWTPCLG-----PTLTGVIAVASATEGSNV
Mflv_3217|M.gilvum_PYR-GCK TFGVLLAVIAQAQATSGWGGLLAVFTAYTAGAATILMLVSVGTAIAGTAL
: : :: . . . :: *
MAB_4275c|M.abscessus_ATCC_199 QATMRKVLPYVGRIAG-VIVLLTGLYVAYYGYYEIRLNF-GDGSADDPVI
MMAR_5058|M.marinum_M TAAFRRILPFVGRIAG-VIVLITGLYVAYYGYYEIRLYF-TSAETDDPVI
Mb2902c|M.bovis_AF2122/97 ADRLRRILPFVNRISG-ALLVVVGLYVGYYGLYELRLIAGVGANPQDAVI
Rv2877c|M.tuberculosis_H37Rv ADRLRRILPFVNRISG-ALLVVVGLYVGYYGLYELRLIAGVGANPQDAVI
MUL_2080|M.ulcerans_Agy99 ADRLRRVLPLVNRISG-GLLAVVGLYVGYYGYYEVRLTSGLLVDPQDRVI
MAV_3728|M.avium_104 ADRLRRMVPFVDRISG-AVLVLVGLYVAYYGGYEVRLFG-AGGDPRDAVI
TH_3805|M.thermoresistible__bu AVRLRRIGRYVNRISG-VLLVAVGGYVAYYGLYELRLFW-GGANPQDPVI
MSMEG_3541|M.smegmatis_MC2_155 IDRMRRILPYANRISG-ALLILVGAYVGYYGLYEVRLFS-GSGDPHDPVI
MLBr_02411|M.leprae_Br4923 TRGILLVIAYCMGLGIPFVLLASGSAQAVAGLRWLRQYG--------RAI
Mvan_0867|M.vanbaalenii_PYR-1 ARGVVLVIAYCLGLGIPFVLLAFGSARAVAGLGWLRRHT--------RTI
Mflv_3217|M.gilvum_PYR-GCK TRHLGILARHGTRVTA-VVLIATGAYLAWYWLPAATGHT-------ASGG
. : : :: * .
MAB_4275c|M.abscessus_ATCC_199 NAAGTVQAWLVGVVDATGVWPLTGGIALIVAVAAASSYAVRKGR------
MMAR_5058|M.marinum_M QAASTLQDSLIRLVDGLSPWLLVGVAAALVAVAVGWHTLARRRASNPAAQ
Mb2902c|M.bovis_AF2122/97 AAAGRLQGALAGWVNQHGAWPWAVLLVVLVVGAFAGTWFR--RVRR----
Rv2877c|M.tuberculosis_H37Rv AAAGRLQGALAGWVNQHGAWPWAVLLVVLVVGAFAGTWFR--RVRR----
MUL_2080|M.ulcerans_Agy99 GAAGRIQGELAGWVHQHGIWPWVVVLTLLVAGAFSVSWYR--RARR----
MAV_3728|M.avium_104 GAAGRLQGALAGWVHRHGLWPWALALLALAAVRFGRARRRGRRADRLPDP
TH_3805|M.thermoresistible__bu EAAARAQGAIAGWVHRHGGGPWLAATAVLMLVVLATGFVA--RIRRSARR
MSMEG_3541|M.smegmatis_MC2_155 AAAGKIQGTLAGWVHQHGALPWAIGLLVLIG--LGLLWRVRIFARRSPAR
MLBr_02411|M.leprae_Br4923 QVFGGMLLIAIGAALIAGVWDDFVSWLRDAVVSDMRVSI-----------
Mvan_0867|M.vanbaalenii_PYR-1 QVLGGVLMILVGAALVTGLWNDFVSWIRDAFVSDVRLPI-----------
Mflv_3217|M.gilvum_PYR-GCK NLLTGWSATATGWLQDHALPASVIAAFAVVVSAVAAYWTTHRRPITGKQR
.
MAB_4275c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_5058|M.marinum_M SLPDPTS-------------------------------------------
Mb2902c|M.bovis_AF2122/97 --------------------------------------------------
Rv2877c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2080|M.ulcerans_Agy99 --------------------------------------------------
MAV_3728|M.avium_104 AAAAAPQVADQHHEDQRRESDQKPVLDVADVVAAQHGQQQPEDDQADDVP
TH_3805|M.thermoresistible__bu --------------------------------------------------
MSMEG_3541|M.smegmatis_MC2_155 RATADDAP------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3217|M.gilvum_PYR-GCK RR------------------------------------------------
MAB_4275c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_5058|M.marinum_M --------------------------------------------------
Mb2902c|M.bovis_AF2122/97 --------------------------------------------------
Rv2877c|M.tuberculosis_H37Rv --------------------------------------------------
MUL_2080|M.ulcerans_Agy99 --------------------------------------------------
MAV_3728|M.avium_104 GPVVDPAPSPRRRRHLDGVDAGEVLDLGTGHGESPSDRIGLSNPDSGTTA
TH_3805|M.thermoresistible__bu --------------------------------------------------
MSMEG_3541|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02411|M.leprae_Br4923 --------------------------------------------------
Mvan_0867|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3217|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4275c|M.abscessus_ATCC_199 -------------------
MMAR_5058|M.marinum_M -------------------
Mb2902c|M.bovis_AF2122/97 -------------------
Rv2877c|M.tuberculosis_H37Rv -------------------
MUL_2080|M.ulcerans_Agy99 -------------------
MAV_3728|M.avium_104 GRARGARPHHGPRRWRAAQ
TH_3805|M.thermoresistible__bu -------------------
MSMEG_3541|M.smegmatis_MC2_155 -------------------
MLBr_02411|M.leprae_Br4923 -------------------
Mvan_0867|M.vanbaalenii_PYR-1 -------------------
Mflv_3217|M.gilvum_PYR-GCK -------------------