For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKAMTYRGPYKVRVEEKDVPSLEHPNDAIVRVQRAAICGSDLHLYHGLMPDTRVGHTFGHEFIGTVHEVG SSVQNLQPGDRVMVPFNIFCGSCYFCARGLYSNCHNVNPNATAVGGIYGYSHTCGGYDGGQAEFVRVPFA DVGPVLIPDWLGDDDALMLTDALPTGYFGAQLGDIVEGDTVIVFGAGPVGLYAAKSAWLMGAGRVLVVDH LDYRLKKAESFAHAETYNFVEYDDIVVEMKKATGYLGADVVIDAVGAEADGNALMHVTSAKLKLQGGSPV ALNWAIDSVRKGGTVSVVGAYGPMFSAVKFGDAMNKGLTINANQCPVKRQWPRLLSHIQQGYLTPSDIVT HHIPLDEIAEGYHMFSSKLDDCIKPVVVMNTN
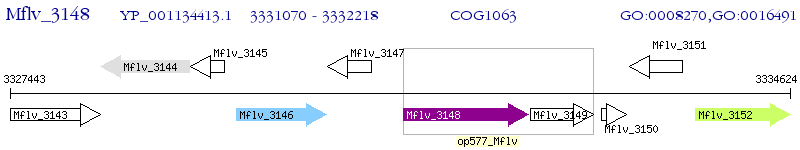
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3148 | - | - | 100% (382) | alcohol dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1966 | - | 1e-58 | 36.90% (393) | alcohol dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4997 | - | 2e-49 | 34.03% (385) | alcohol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1929 | - | 3e-29 | 34.87% (238) | dehydrogenase |
| M. tuberculosis H37Rv | Rv1895 | - | 3e-46 | 34.47% (380) | dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 1e-22 | 28.70% (345) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_1869c | - | 2e-25 | 27.70% (361) | zinc-dependent alcohol dehydrogenase AdhE2 |
| M. marinum M | MMAR_2790 | - | 5e-47 | 35.11% (376) | dehydrogenase |
| M. avium 104 | MAV_4101 | - | 1e-57 | 36.13% (393) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0671 | - | 0.0 | 82.28% (378) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. thermoresistible (build 8) | TH_2319 | - | 2e-45 | 40.08% (242) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. ulcerans Agy99 | MUL_2962 | - | 2e-44 | 34.31% (376) | dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4749 | - | 6e-60 | 36.68% (398) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1929|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv -----MRAVVIDGAGSVRVNTQPDPALPGPDGVVVAVTAAGICGSDLHFY
MMAR_2790|M.marinum_M -----MRTVVIDGPGSIRVDNRPDPALPGSDGVIVAVSAAGICGSDLHFY
MUL_2962|M.ulcerans_Agy99 -----MRTVVIDGPGSIRVDNRPDPALPGSDGVIVVVSAAGICGSDLHFY
Mflv_3148|M.gilvum_PYR-GCK -----MKAMTYRGPYKVRVEEKDVPSLEHPNDAIVRVQRAAICGSDLHLY
MSMEG_0671|M.smegmatis_MC2_155 -----MRAMVYRGPYKIRLEDKPIPAIEHPNDAIVRVTKAAICGSDLHLY
MAV_4101|M.avium_104 -----MKAVTWHGKRDVRVDSVPDPKIEQPTDAIIEVTSTNICGSDLHLY
Mvan_4749|M.vanbaalenii_PYR-1 -----MRAVTWHGRRDVRVDSVPDPKIEEPTDAIIEVTSTNICGSDLHLY
TH_2319|M.thermoresistible__bu --------VTWHGRRKVSVDTVPDPAIMEPTDAIIRVTSTNICGSDLHLY
MLBr_01784|M.leprae_Br4923 MSQTVRGVISRKKDEPVELVDIVIPDP-GPGEAVVDVTACGVCHTDLTYR
MAB_1869c|M.abscessus_ATCC_199 ----------------MELVDIVIPDP-GPGEVVVKVQACGVCHTDLTYR
Mb1929|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv EGEYPFTEP-VALGHEAVGTIVEAGPQVRTVGVGDLVMVSSVAGCGVCPG
MMAR_2790|M.marinum_M EGEYPLAEP-VALGHEAVGTVVEKGPDVHTVEVGDQVMVSSVAGCGACAG
MUL_2962|M.ulcerans_Agy99 EGEYPLAEP-VALGHEAVGTVVEKGPDVHTVEVGDQVMVSSVAGCGACAG
Mflv_3148|M.gilvum_PYR-GCK HGLMPDTRVGHTFGHEFIGTVHEVGSSVQNLQPGDRVMVPFNIFCGSCYF
MSMEG_0671|M.smegmatis_MC2_155 HGMMPDTRVGTTFGHEFIGVVEKVGPSVQNLSVGDRVMVPFNIFCGSCYF
MAV_4101|M.avium_104 EILGAFMKPGDILGHEPMGVVREVGTETGDLRVGDRVVIPFQICCGSCYM
Mvan_4749|M.vanbaalenii_PYR-1 EVLGAFMNEGDILGHEPMGVVREVGSAVSNLAVGDRVVIPFQISCGHCFM
TH_2319|M.thermoresistible__bu EVLSAFMSPGDILGHEAMGVVEEVGPEVGALQAGDRVVIPFNISCGTCWM
MLBr_01784|M.leprae_Br4923 EGGINDRYP-FLLGHEAAGTVEVVGPGVTAVEPGDFVILNWRAVCGQCRA
MAB_1869c|M.abscessus_ATCC_199 EGGINDEFP-FLLGHEAAGIVETVGEGVTTVEPGDFVILNWRAVCGVCRA
Mb1929|M.bovis_AF2122/97 -------------M---IFGA---------GVLGGAQADLLAVPAADFQV
Rv1895|M.tuberculosis_H37Rv CETHDPVMCFSGPM---IFGA---------GVLGGAQADLLAVPAADFQV
MMAR_2790|M.marinum_M CATRDPAMCVSGPQ---IFGS---------GALGGAQADLLAVPAADFQV
MUL_2962|M.ulcerans_Agy99 CATRDPAMCVSGPQ---IFGS---------GALGGAQADLLAVPATDFQV
Mflv_3148|M.gilvum_PYR-GCK CARGLYSNCHNVNPNATAVGG-IYGYSHTCGGYDGGQAEFVRVPFADVGP
MSMEG_0671|M.smegmatis_MC2_155 CARGLYSNCHNVNPNATAVGG-IYGYSHTCGGYDGGQAEFVRVPFADVGP
MAV_4101|M.avium_104 CGQQLYTQCETTQVRDQGMGAALFGYSELYGEVPGGQAELLRVPQAQFTH
Mvan_4749|M.vanbaalenii_PYR-1 CDRKLYTQCETTQVRDQGMGAALFGYSELYGSVPGGQAEYLRVPQAQFTH
TH_2319|M.thermoresistible__bu CAQGLQSQCETTQNRDQGTGAALFGYSKLYGEVAGGQAEYLRVPQAQYTH
MLBr_01784|M.leprae_Br4923 CKRGRPSYCFDTFN--AQQKMTLIDGTELTPALGIGAFADKTLVHSGQCT
MAB_1869c|M.abscessus_ATCC_199 CKRGRPQYCFDTFN--AAQKMTLTDGTELTPALGIGAFIEKTLVHAGQCT
. : :
Mb1929|M.bovis_AF2122/97 LKIPEGITTEQALLLTDNLATGWAAAQRADISFGSAVAVIGLGAVGLCAL
Rv1895|M.tuberculosis_H37Rv LKIPEGITTEQALLLTDNLATGWAAAQRADISFGSAVAVIGLGAVGLCAL
MMAR_2790|M.marinum_M LKIPAAITTEQALLLTDNLATGWAAARRADILFGATVAVIGLGAVGLCAV
MUL_2962|M.ulcerans_Agy99 LKIPAAITTEQALLLTDNLATGWAAARRADIPFGATVAVIGLGAVGLCAV
Mflv_3148|M.gilvum_PYR-GCK VLIPDWLGDDDALMLTDALPTGYFGAQLGDIVEGDTVIVFGAGPVGLYAA
MSMEG_0671|M.smegmatis_MC2_155 SIIPDRLDDEDALLLTDALATGYFGAQLGNIAEGDTVVVFGAGPVGLFAA
MAV_4101|M.avium_104 IKVPVGPPDSRFVYLSDVLPTAWQAVAYADIPDGGTVAVLGLGPIGDMAS
Mvan_4749|M.vanbaalenii_PYR-1 IKVPDGPPDSRFVYLSDVLPTAWQAVAYADVPDGGTVTVLGLGPIGDMAA
TH_2319|M.thermoresistible__bu ITVPHDGPDDRYVYLSDVLPTAWQGVEYAAVPDGGTLVVLGLGPIGSMAC
MLBr_01784|M.leprae_Br4923 KVDPAADPAVAGLLGCGVMAGLGAAINTAAVSRDDTVAVIGCGGVGDAAI
MAB_1869c|M.abscessus_ATCC_199 KVDPTADPAVVGLLGCGVMAGLGAAVNTGNVGRDDTVAVIGCGGVGDAAI
* : . :. . . : . :: *:* * :* *
Mb1929|M.bovis_AF2122/97 RSAFIHGAATVFAVDRVKGRLQRAATWG---------ATPIPSPAAETIL
Rv1895|M.tuberculosis_H37Rv RSAFIHGAATVFAVDRVKGRLQRAATWG---------ATPIPSPAAETIL
MMAR_2790|M.marinum_M RSALAQGAATVFAVDRVQGRLQRASEWG---------ATAVASPAAEAIV
MUL_2962|M.ulcerans_Agy99 RSALAQGAATVFAVDRVQGRLQRAAEWG---------ATAVASPAAEAIV
Mflv_3148|M.gilvum_PYR-GCK KSAWLMGAGRVLVVDHLDYRLKKAESFAHAETYNFVEYDDIVVEMKKATG
MSMEG_0671|M.smegmatis_MC2_155 KSAWLMGAGRVIVIDHLEYRLEKARTFAHAETINFVDCDDIVVKMKRTTD
MAV_4101|M.avium_104 RIADHLG-YRVIAVDLVPERLG--RAAQRGIHT--IDLARLDAGLGDAIR
Mvan_4749|M.vanbaalenii_PYR-1 RIAQHLG-YQVFAVDLVPERLD--RAIARGIHT--IDASIVDGSVGDEVR
TH_2319|M.thermoresistible__bu RIAAHRGNCRVIGVDRVPERIEKVRPYCADVLN--VDTDDVEAAVREQTG
MLBr_01784|M.leprae_Br4923 SGAALVGANRIIAVDIDDTKLEWARTFGATHTVN----ALEL-EVVKTIQ
MAB_1869c|M.abscessus_ATCC_199 AGARLAGARKIIAIDTDDTKLAWAKDFGATDTIN----ARKVDDVVTAIQ
* * :: :* ::
Mb1929|M.bovis_AF2122/97 AATRGRGADSVIDAVGTDASMSDALN------------------------
Rv1895|M.tuberculosis_H37Rv AATRGRGADSVIDAVGTDASMSDALN------------------------
MMAR_2790|M.marinum_M AATGGRGADSVIDAVGTDASMTDALN------------------------
MUL_2962|M.ulcerans_Agy99 AATSGRGADSVIDAVDTDASMTDALN------------------------
Mflv_3148|M.gilvum_PYR-GCK YLG----ADVVIDAVGAEADGNALMHVTSAKLKLQ---------------
MSMEG_0671|M.smegmatis_MC2_155 HLG----ADVAIDAAGAEADGNFLQHVTATKLKLQ---------------
MAV_4101|M.avium_104 DLTDGRGADSVIDAVGMEAHGSPAAKLAQQVAGVLPDALGKR------LM
Mvan_4749|M.vanbaalenii_PYR-1 RLTGGRGSDSVIDAVGMEAHGSPVAKFAQQATALLPDAVAKP------MM
TH_2319|M.thermoresistible__bu GXXVQTPPDPRRQSADCENHADQHGRPEHPPEELHHQADDQRQRRHERDA
MLBr_01784|M.leprae_Br4923 DLTSGFGVDVVIDTVGRPETWKQAFY------------------------
MAB_1869c|M.abscessus_ATCC_199 ELTDEFGADVVIDAVGRPETWKQAFY------------------------
* ::..
Mb1929|M.bovis_AF2122/97 ---------------------AVRPGGTVSVVG-----------------
Rv1895|M.tuberculosis_H37Rv ---------------------AVRPGGTVSVVG-----------------
MMAR_2790|M.marinum_M ---------------------AVRAGGTVSVVG-----------------
MUL_2962|M.ulcerans_Agy99 ---------------------AVRAGGTVSVVG-----------------
Mflv_3148|M.gilvum_PYR-GCK ---GGSPVALNW------AIDSVRKGGTVSVVG-------------AYG-
MSMEG_0671|M.smegmatis_MC2_155 ---GGSPIALNW------AIDSVRKGGTVSVVG-------------AYG-
MAV_4101|M.avium_104 QTAGVDRLSALY-----SAIDAVRRGGTISLIG-------------VYG-
Mvan_4749|M.vanbaalenii_PYR-1 QKAGVDRLDALY-----TAIDCVRRGGTLSLIG-------------VYG-
TH_2319|M.thermoresistible__bu ATAGSPPLPSPWPDRTTTHRHTWRRLGGMELLRDVVVLLHIVGFAVTFGA
MLBr_01784|M.leprae_Br4923 ---------------------ARDLAGTVVLVG-----------------
MAB_1869c|M.abscessus_ATCC_199 ---------------------ARDLAGTVVLVG-----------------
* : ::
Mb1929|M.bovis_AF2122/97 ---VHDLQPFPLPALTCLLRSITLRMT-MAPVQRTWPELIP---------
Rv1895|M.tuberculosis_H37Rv ---VHDLQPFPVPALTCLLRSITLRMT-MAPVQRTWPELIP---------
MMAR_2790|M.marinum_M ---VHDLQPFPLPALTCLLRSITLRLT-IAPVQRTWSELIP---------
MUL_2962|M.ulcerans_Agy99 ---VHDLQPFPLPALTCLLRSITLRLT-IAPVQRTWSELIP---------
Mflv_3148|M.gilvum_PYR-GCK ----PMFSAVKFGDAMNKG--LTINAN-QCPVKRQWPRLLS---------
MSMEG_0671|M.smegmatis_MC2_155 ----PMFSAVKFGDAFNKG--LTLRMN-QCPVKREWPRLLS---------
MAV_4101|M.avium_104 ----GMADPLPMLTLFDK--QVTLRMG-QANVKRWVDDIMP---------
Mvan_4749|M.vanbaalenii_PYR-1 ----GMADPMPMLTLFDK--QIQVRMG-QANVKKWVDDIMP---------
TH_2319|M.thermoresistible__bu WTAEAVARRFRTTRLMDYGLLLSLITG-LALAAPWPAGIVLNYPKIGLKL
MLBr_01784|M.leprae_Br4923 VPTPDMRLDMPLLDFFSHGGSLKSSWYGDCLPERDFPTLVD---------
MAB_1869c|M.abscessus_ATCC_199 VPTPDMTLEMPLIDFFSRGGSLKSSWYGDCLPERDFPTLVS---------
. : . ::
Mb1929|M.bovis_AF2122/97 --LLQSGRLDVDGIFTTTLPLDEAAKGYATARARSGEELKVLLTP-----
Rv1895|M.tuberculosis_H37Rv --LLQSGRLDVDGIFTTTLPLDEAAKGYATARARSGEELRFCLRPDSRDV
MMAR_2790|M.marinum_M --LLESGRLDVDGIFTTSLPLDEAAKGYSIAGSRSGNDVKVLLKV-----
MUL_2962|M.ulcerans_Agy99 --LLESGRLDVDGIFTTSLPLDEAAKGYSIAGSRSGNDVKVLLKV-----
Mflv_3148|M.gilvum_PYR-GCK --HIQQGYLTPSDIVTHHIPLDEIAEGYHMFSSKLDDCIKPVVVMNTN--
MSMEG_0671|M.smegmatis_MC2_155 --HIKNGYLKPNDIVTHRIPLEHIAEGYHIFSAKLDDCIKPIIVPTAS--
MAV_4101|M.avium_104 --LLTDDDPLGVDDFATHVL--PLERAPHAYEIFQKKQDGAVKVILKP--
Mvan_4749|M.vanbaalenii_PYR-1 --LLTDSDPLGVDTFATHVL--PMEEAPHAYKIFQQKQDGAVKVILQP--
TH_2319|M.thermoresistible__bu VLLVALGAVLGIGGAHQRRTGNPVPRPLFVTAGVLSFAAAAVAVLW----
MLBr_01784|M.leprae_Br4923 --LYLQGRLPLGKFVSERIGLGDVEEAFHKIHG--GKVLRSVVML-----
MAB_1869c|M.abscessus_ATCC_199 --LYQQGRLPLDRFVSERIGIESIEAAFEKMHH--GEVLRSVVVL-----
.
Mb1929|M.bovis_AF2122/97 --------------------------------------
Rv1895|M.tuberculosis_H37Rv LGAHETVDLYVHVRRCQSVADLQLEGAADGVDGPSMLN
MMAR_2790|M.marinum_M --------------------------------------
MUL_2962|M.ulcerans_Agy99 --------------------------------------
Mflv_3148|M.gilvum_PYR-GCK --------------------------------------
MSMEG_0671|M.smegmatis_MC2_155 --------------------------------------
MAV_4101|M.avium_104 --------------------------------------
Mvan_4749|M.vanbaalenii_PYR-1 --------------------------------------
TH_2319|M.thermoresistible__bu --------------------------------------
MLBr_01784|M.leprae_Br4923 --------------------------------------
MAB_1869c|M.abscessus_ATCC_199 --------------------------------------