For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RAVTWHGRRDVRVDSVPDPKIEEPTDAIIEVTSTNICGSDLHLYEVLGAFMNEGDILGHEPMGVVREVGS AVSNLAVGDRVVIPFQISCGHCFMCDRKLYTQCETTQVRDQGMGAALFGYSELYGSVPGGQAEYLRVPQA QFTHIKVPDGPPDSRFVYLSDVLPTAWQAVAYADVPDGGTVTVLGLGPIGDMAARIAQHLGYQVFAVDLV PERLDRAIARGIHTIDASIVDGSVGDEVRRLTGGRGSDSVIDAVGMEAHGSPVAKFAQQATALLPDAVAK PMMQKAGVDRLDALYTAIDCVRRGGTLSLIGVYGGMADPMPMLTLFDKQIQVRMGQANVKKWVDDIMPLL TDSDPLGVDTFATHVLPMEEAPHAYKIFQQKQDGAVKVILQP*
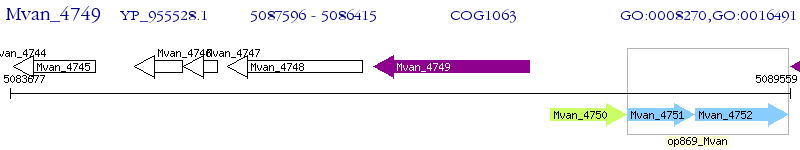
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4749 | - | - | 100% (393) | alcohol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1841 | - | 1e-52 | 35.95% (395) | alcohol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4901 | - | 6e-42 | 32.15% (395) | alcohol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1929 | - | 2e-30 | 33.83% (269) | dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1966 | - | 0.0 | 88.04% (393) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv1895 | - | 1e-48 | 34.01% (394) | dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 4e-18 | 29.50% (322) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_2084 | - | 4e-15 | 31.85% (248) | zinc-type alcohol dehydrogenase AdhD |
| M. marinum M | MMAR_2790 | - | 1e-48 | 34.77% (394) | dehydrogenase |
| M. avium 104 | MAV_4101 | - | 0.0 | 82.44% (393) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6616 | - | 0.0 | 83.97% (393) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. thermoresistible (build 8) | TH_2319 | - | 4e-90 | 68.57% (245) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. ulcerans Agy99 | MUL_2962 | - | 5e-48 | 34.26% (394) | dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1929|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv ----MRAVVIDGAGSVRVNTQPDPALPGPDGVVVAVTAAGICGSDLHFYE
MMAR_2790|M.marinum_M ----MRTVVIDGPGSIRVDNRPDPALPGSDGVIVAVSAAGICGSDLHFYE
MUL_2962|M.ulcerans_Agy99 ----MRTVVIDGPGSIRVDNRPDPALPGSDGVIVVVSAAGICGSDLHFYE
Mvan_4749|M.vanbaalenii_PYR-1 ----MRAVTWHGRRDVRVDSVPDPKIEEPTDAIIEVTSTNICGSDLHLYE
Mflv_1966|M.gilvum_PYR-GCK ----MKAVTWHGRRDVRVDTVADPKIEQPTDAIIEVTSTNICGSDLHLYE
MSMEG_6616|M.smegmatis_MC2_155 ----MKAVTWHGRRDVRVDNVPDPKIEQPTDAIIEVTSTNICGSDLHLYE
MAV_4101|M.avium_104 ----MKAVTWHGKRDVRVDSVPDPKIEQPTDAIIEVTSTNICGSDLHLYE
TH_2319|M.thermoresistible__bu -------VTWHGRRKVSVDTVPDPAIMEPTDAIIRVTSTNICGSDLHLYE
MLBr_01784|M.leprae_Br4923 MSQTVRGVISRKKDEPVELVDIVIPDPGPGEAVVDVTACGVCHTDLTYRE
MAB_2084|M.abscessus_ATCC_1997 --MKTRAAVLWGLEQKWEVEEVDLDPPGPGEVLVRLAATGLCHSDEHLVT
Mb1929|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv GEYPFTEP-VALGHEAVGTIVEAGPQVRTVGVGDLVMVSSVAGCGVCPGC
MMAR_2790|M.marinum_M GEYPLAEP-VALGHEAVGTVVEKGPDVHTVEVGDQVMVSSVAGCGACAGC
MUL_2962|M.ulcerans_Agy99 GEYPLAEP-VALGHEAVGTVVEKGPDVHTVEVGDQVMVSSVAGCGACAGC
Mvan_4749|M.vanbaalenii_PYR-1 VLGAFMNEGDILGHEPMGVVREVGSAVSNLAVGDRVVIPFQISCGHCFMC
Mflv_1966|M.gilvum_PYR-GCK VLGAFMSEGDVLGHEPMGIVREVGAEVSNLSVGDRVVIPFQISCGHCFMC
MSMEG_6616|M.smegmatis_MC2_155 VLGAFMNEGDILGHEPMGIVREVGSAVDGLSVGDRVVIPFQISCGHCFMC
MAV_4101|M.avium_104 ILGAFMKPGDILGHEPMGVVREVGTETGDLRVGDRVVIPFQICCGSCYMC
TH_2319|M.thermoresistible__bu VLSAFMSPGDILGHEAMGVVEEVGPEVGALQAGDRVVIPFNISCGTCWMC
MLBr_01784|M.leprae_Br4923 GGINDRYP-FLLGHEAAGTVEVVGPGVTAVEPGDFVILNWRAVCGQCRAC
MAB_2084|M.abscessus_ATCC_1997 GDLPIPLP-VVGGHEGAGTVVECGEGVEDLSEGDSVILTFLPSCGRCSYC
Mb1929|M.bovis_AF2122/97 --------------------MIFG----AGVLGG-----AQADLLAVPAA
Rv1895|M.tuberculosis_H37Rv ETHDPVMCFS--------GPMIFG----AGVLGG-----AQADLLAVPAA
MMAR_2790|M.marinum_M ATRDPAMCVS--------GPQIFG----SGALGG-----AQADLLAVPAA
MUL_2962|M.ulcerans_Agy99 ATRDPAMCVS--------GPQIFG----SGALGG-----AQADLLAVPAT
Mvan_4749|M.vanbaalenii_PYR-1 DRKLYTQCETTQVRDQGMGAALFGYSELYGSVPG-----GQAEYLRVPQA
Mflv_1966|M.gilvum_PYR-GCK DMKLYTQCETTQVREQGMGAALFGFSELYGSVPG-----GQAEYLRVPQA
MSMEG_6616|M.smegmatis_MC2_155 DQKLYTQCETTQVREQGMGAALFGYSELYGSVPG-----GQAQYLRVPQA
MAV_4101|M.avium_104 GQQLYTQCETTQVRDQGMGAALFGYSELYGEVPG-----GQAELLRVPQA
TH_2319|M.thermoresistible__bu AQGLQSQCETTQNRDQGTGAALFGYSKLYGEVAG-----GQAEYLRVPQA
MLBr_01784|M.leprae_Br4923 KRGRPSYCFDTFN--AQQKMTLIDGTELTPALGI-----GAFADKTLVHS
MAB_2084|M.abscessus_ATCC_1997 ARGMGNLCELGAALMMGPQIDGTYRFHARGEDVGQMCLLGTFSEYTVVPQ
. :
Mb1929|M.bovis_AF2122/97 DFQVLKIPEGITTEQALLLTDNLATGWAAAQRADISFGSAVAVIGLGAVG
Rv1895|M.tuberculosis_H37Rv DFQVLKIPEGITTEQALLLTDNLATGWAAAQRADISFGSAVAVIGLGAVG
MMAR_2790|M.marinum_M DFQVLKIPAAITTEQALLLTDNLATGWAAARRADILFGATVAVIGLGAVG
MUL_2962|M.ulcerans_Agy99 DFQVLKIPAAITTEQALLLTDNLATGWAAARRADIPFGATVAVIGLGAVG
Mvan_4749|M.vanbaalenii_PYR-1 QFTHIKVPDGPPDSRFVYLSDVLPTAWQAVAYADVPDGGTVTVLGLGPIG
Mflv_1966|M.gilvum_PYR-GCK QFTHIKVPEGPDDSRFVYLSDVLPTAWQSVAYADIPDGGTVTVIGLGPIG
MSMEG_6616|M.smegmatis_MC2_155 QFTHIKVPDGPPDSRYVYLSDVLPTAWQSVAYAEIPKGGTVTVLGLGPIG
MAV_4101|M.avium_104 QFTHIKVPVGPPDSRFVYLSDVLPTAWQAVAYADIPDGGTVAVLGLGPIG
TH_2319|M.thermoresistible__bu QYTHITVPHDGPDDRYVYLSDVLPTAWQGVEYAAVPDGGTLVVLGLGPIG
MLBr_01784|M.leprae_Br4923 GQCTKVDPAADPAVAGLLGCGVMAGLGAAINTAAVSRDDTVAVIGCGGVG
MAB_2084|M.abscessus_ATCC_1997 ASVVKIDGGIPLDRAALIGCGVTTGYGSAVRTGEVRAGDTVVVVGAGGIG
: . . . . : . ::.*:* * :*
Mb1929|M.bovis_AF2122/97 LCALRSAFIHGAATVFAVDRVKGRLQ--RAATWGATPIPS-----PAAET
Rv1895|M.tuberculosis_H37Rv LCALRSAFIHGAATVFAVDRVKGRLQ--RAATWGATPIPS-----PAAET
MMAR_2790|M.marinum_M LCAVRSALAQGAATVFAVDRVQGRLQ--RASEWGATAVAS-----PAAEA
MUL_2962|M.ulcerans_Agy99 LCAVRSALAQGAATVFAVDRVQGRLQ--RAAEWGATAVAS-----PAAEA
Mvan_4749|M.vanbaalenii_PYR-1 DMAARIAQHLG-YQVFAVDLVPERLD--RAIARGIHTIDASIVDGSVGDE
Mflv_1966|M.gilvum_PYR-GCK DMAARIAQHKG-HTVFAVDFVPERLA--RAKARGIHTIDASALDGSVGDE
MSMEG_6616|M.smegmatis_MC2_155 DMAARIAHHLG-YEVFAVDRIPERLR--RAADRGIQAIDLGAIDEPLGDV
MAV_4101|M.avium_104 DMASRIADHLG-YRVIAVDLVPERLG--RAAQRGIHTIDLARLDAGLGDA
TH_2319|M.thermoresistible__bu SMACRIAAHRGNCRVIGVDRVPERIEKVRPYCADVLNVDTDDVEAAVREQ
MLBr_01784|M.leprae_Br4923 DAAISGAALVGANRIIAVDIDDTKLE--WARTFGATHTVN-ALELEVVKT
MAB_2084|M.abscessus_ATCC_1997 MNAIQGARIAGALAIVAVDPVNFKRE--QASGFGATHTAAS--MDEAWSL
* * * :..** : . . .
Mb1929|M.bovis_AF2122/97 ILAATRGRGADSVIDAVGTDASMS--------------------------
Rv1895|M.tuberculosis_H37Rv ILAATRGRGADSVIDAVGTDASMS--------------------------
MMAR_2790|M.marinum_M IVAATGGRGADSVIDAVGTDASMT--------------------------
MUL_2962|M.ulcerans_Agy99 IVAATSGRGADSVIDAVDTDASMT--------------------------
Mvan_4749|M.vanbaalenii_PYR-1 VRRLTGGRGSDSVIDAVGMEAHGSPVAKFAQQATALLPDAVAKP------
Mflv_1966|M.gilvum_PYR-GCK VRRLTSGRGSDSVIDAVGMEAHGSPVAKAVQDATAFLPGAIAKP------
MSMEG_6616|M.smegmatis_MC2_155 IRSMTHGRGTDAVIDAVGMEAHGSPVAKIMQTAAAALPDAVAKP------
MAV_4101|M.avium_104 IRDLTDGRGADSVIDAVGMEAHGSPAAKLAQQVAGVLPDALGKR------
TH_2319|M.thermoresistible__bu TGGXXVQTPPDPRRQSADCENHADQHGRPEHPPEELHHQADDQRQRRHER
MLBr_01784|M.leprae_Br4923 IQDLTSGFGVDVVIDTVGRPETWK--------------------------
MAB_2084|M.abscessus_ATCC_1997 VSDMTRGKLADVCILTTDVAEGSYT-------------------------
* :..
Mb1929|M.bovis_AF2122/97 --------------DALNAVRPGGTVSVVGVHD--LQPFPLPALTCLLRS
Rv1895|M.tuberculosis_H37Rv --------------DALNAVRPGGTVSVVGVHD--LQPFPVPALTCLLRS
MMAR_2790|M.marinum_M --------------DALNAVRAGGTVSVVGVHD--LQPFPLPALTCLLRS
MUL_2962|M.ulcerans_Agy99 --------------DALNAVRAGGTVSVVGVHD--LQPFPLPALTCLLRS
Mvan_4749|M.vanbaalenii_PYR-1 MMQKAGVDRLDALYTAIDCVRRGGTLSLIGVYG--GMADPMPMLTLFDKQ
Mflv_1966|M.gilvum_PYR-GCK MMKKAGVDRLDALYTAIDCVRRGGTLSVIGVYG--GMADPMPILTMFDKQ
MSMEG_6616|M.smegmatis_MC2_155 MMENAGVDRLDALYSAIDTVRRGGTISLIGVYG--GTADPIPMLTLFDKQ
MAV_4101|M.avium_104 LMQTAGVDRLSALYSAIDAVRRGGTISLIGVYG--GMADPLPMLTLFDKQ
TH_2319|M.thermoresistible__bu DAATAGSPPLPSPWPDRTTTHR-HTWRRLGGME--LLRDVVVLLHIVGFA
MLBr_01784|M.leprae_Br4923 --------------QAFYARDLAGTVVLVGVPTP-DMRLDMPLLDFFSHG
MAB_2084|M.abscessus_ATCC_1997 -------------AEALALVGKRGRVVVTAIGHPDESSISGSLLEMTLYE
. *
Mb1929|M.bovis_AF2122/97 ITL---RMTMAPVQRTWPE-------LIPLLQ----------SGRLDVDG
Rv1895|M.tuberculosis_H37Rv ITL---RMTMAPVQRTWPE-------LIPLLQ----------SGRLDVDG
MMAR_2790|M.marinum_M ITL---RLTIAPVQRTWSE-------LIPLLE----------SGRLDVDG
MUL_2962|M.ulcerans_Agy99 ITL---RLTIAPVQRTWSE-------LIPLLE----------SGRLDVDG
Mvan_4749|M.vanbaalenii_PYR-1 IQV---RMGQANVKKWVDD-------IMPLLT---------DSDPLGVDT
Mflv_1966|M.gilvum_PYR-GCK IQV---RMGQANVKRWVDD-------ILPLLT---------DEDPLGVDT
MSMEG_6616|M.smegmatis_MC2_155 IQL---RMGQANVKRWVDD-------IMPLLG---------DDDPLGVDD
MAV_4101|M.avium_104 VTL---RMGQANVKRWVDD-------IMPLLT---------DDDPLGVDD
TH_2319|M.thermoresistible__bu VTFGA-WTAEAVARRFRTTRLMDYGLLLSLITGLALAAPWPAGIVLNYPK
MLBr_01784|M.leprae_Br4923 GSLKSSWYGDCLPERDFPT-------LVDLYL----------QGRLPLGK
MAB_2084|M.abscessus_ATCC_1997 KQIRGALYGSSNAPHDIPR-------LVELYS----------AGLLKLDE
. . : :: * *
Mb1929|M.bovis_AF2122/97 IFTTTLPLDEAAKGYATARARSGEELKVLLTP------------------
Rv1895|M.tuberculosis_H37Rv IFTTTLPLDEAAKGYATARARSGEELRFCLRPDSRDVLGAHETVDLYVHV
MMAR_2790|M.marinum_M IFTTSLPLDEAAKGYSIAGSRSGNDVKVLLKV------------------
MUL_2962|M.ulcerans_Agy99 IFTTSLPLDEAAKGYSIAGSRSGNDVKVLLKV------------------
Mvan_4749|M.vanbaalenii_PYR-1 FATHVLPMEEAPHAYKIFQQKQDGAVKVILQP------------------
Mflv_1966|M.gilvum_PYR-GCK FATHTLPLDQAPHAYEIFQKKQDGAVKVILKP------------------
MSMEG_6616|M.smegmatis_MC2_155 FATHILPIDEAPHAYEIFQKKQDGAVKIILKP------------------
MAV_4101|M.avium_104 FATHVLPLERAPHAYEIFQKKQDGAVKVILKP------------------
TH_2319|M.thermoresistible__bu IGLKLVLLVALGAVLGIGGAHQRRTGNPVPRPLFVTAGVLSFAAAAVAVL
MLBr_01784|M.leprae_Br4923 FVSERIGLGDVEEAFHKIHGGKVLRSVVML--------------------
MAB_2084|M.abscessus_ATCC_1997 LVTREYSLDQINEGYQDMRSGRNIRGLVRF--------------------
: :
Mb1929|M.bovis_AF2122/97 -------------------------
Rv1895|M.tuberculosis_H37Rv RRCQSVADLQLEGAADGVDGPSMLN
MMAR_2790|M.marinum_M -------------------------
MUL_2962|M.ulcerans_Agy99 -------------------------
Mvan_4749|M.vanbaalenii_PYR-1 -------------------------
Mflv_1966|M.gilvum_PYR-GCK -------------------------
MSMEG_6616|M.smegmatis_MC2_155 -------------------------
MAV_4101|M.avium_104 -------------------------
TH_2319|M.thermoresistible__bu W------------------------
MLBr_01784|M.leprae_Br4923 -------------------------
MAB_2084|M.abscessus_ATCC_1997 -------------------------