For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RAKDLVDARYAEPITVDDLAAAAGLSRAHFSRMFTRTFGESPRAYLQTRRLERAASLLRHTDRSVADICV MVGLQSVGSFTTSFARAYGLPPAAYRASMPSAAVHARIPSCILQRDTRPKADSRVKTAHGEKTGANASP*
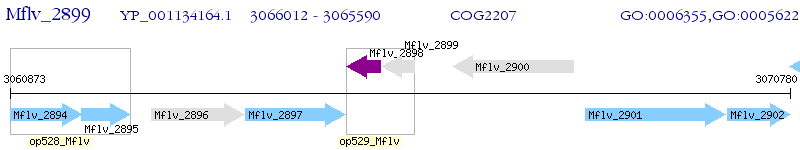
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2899 | - | - | 100% (140) | helix-turn-helix domain-containing protein |
| M. gilvum PYR-GCK | Mflv_5242 | - | 3e-12 | 38.89% (90) | helix-turn-helix domain-containing protein |
| M. gilvum PYR-GCK | Mflv_1693 | - | 1e-10 | 33.33% (87) | AraC family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1966c | - | 4e-08 | 33.33% (81) | transcriptional regulatory protein |
| M. tuberculosis H37Rv | Rv1931c | - | 4e-08 | 33.33% (81) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4709c | - | 6e-27 | 46.22% (119) | AraC family transcriptional regulator |
| M. marinum M | MMAR_1063 | - | 5e-13 | 32.99% (97) | transcriptional regulatory protein |
| M. avium 104 | MAV_4981 | - | 7e-07 | 31.63% (98) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_0213 | - | 8e-27 | 46.22% (119) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_2340 | - | 9e-09 | 32.18% (87) | transcriptional regulatory protein |
| M. ulcerans Agy99 | MUL_0821 | - | 3e-13 | 32.99% (97) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3612 | - | 6e-69 | 94.12% (136) | helix-turn-helix domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3612|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0213|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1063|M.marinum_M ----------------MAQAARLIQHRSGVPIYGYRTDPDIPRVSVLRPG
MUL_0821|M.ulcerans_Agy99 ----------MDYLWRMAQAARLIQHRSGVPIYGYRTDPDIPRVSVRRPG
Mb1966c|M.bovis_AF2122/97 -----------------------------MVIVGFPGDPVDTVILPGGAG
Rv1931c|M.tuberculosis_H37Rv -----------------------------MVIVGFPGDPVDTVILPGGAG
MAV_4981|M.avium_104 MADSSATPRVVVIVVFDDVTMLDVAGAGEVFAEANRFGADYLLKIASVDG
TH_2340|M.thermoresistible__bu ------------------------VTVSALSARNLASSAKPPQIDLRRGG
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3612|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0213|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1063|M.marinum_M PEDLGDRARHIHDFPALWYVPAAGLVYIAAAGQVLEPTQFGSLQGGVAMF
MUL_0821|M.ulcerans_Agy99 PEDLGDRARHIHDFPALWYVPAAGLVYIAAAGQVLEPTQFGSLQAGVAMF
Mb1966c|M.bovis_AF2122/97 VDAARSEPALIDWVKAVSGTARRVVTVCTGAFLAAEAGLLGRTPSDDALG
Rv1931c|M.tuberculosis_H37Rv VDAARSEPALIDWVKAVSGTARRVVTVCTGAFLAAEAGLLGRTPSDDALG
MAV_4981|M.avium_104 RDVTTSIGTRLGVTDAVSSIESADTVLVAGSDNLPRRPIDPELVEAVKSV
TH_2340|M.thermoresistible__bu RALGGSYLYEGDKLQTGWHSHDVHQIEYAIGGVVEVQTRTGHYLLPPQQA
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3612|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_0213|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1063|M.marinum_M FDPAALGGDGGSPWPSWRTHPLLFPFLHGHSGGILRLEVPAARRSTWDGA
MUL_0821|M.ulcerans_Agy99 FDPAALGGDGGSPWPSWRTHPLLFPFLHGHSGGILRLEVPAARRSTWDGA
Mb1966c|M.bovis_AF2122/97 LCRTFRPRISGRSGRCRPDLHAQFAEGVDRGWSHRRHRPRAG--------
Rv1931c|M.tuberculosis_H37Rv LCRTFRPRISGRSGRCRPDLHAQFAEGVDRGWSHRRHRPRAG--------
MAV_4981|M.avium_104 ADRTRRLASICTGSFILGQAGLLDGRRATTHWHDVRLFARAFPEITVEPD
TH_2340|M.thermoresistible__bu AWLPVGLEHSATMNPDVKTVAVMFDPQLVPDAGNRARILAVSPLIR----
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3612|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 ------------------------MLFGTNLGTVTSAGE-----------
MSMEG_0213|M.smegmatis_MC2_155 -----------------------------------MPGE-----------
MMAR_1063|M.marinum_M IAAIEAELTARQQGYGQAVAAHLTLLLIELARLARDVVG-----------
MUL_0821|M.ulcerans_Agy99 IAAIEAELTTRQQGYGQAVTAHLTLLLIELARLARDVVG-----------
Mb1966c|M.bovis_AF2122/97 ----TGRRRPRHRDCPDGCPLARPVSAPTRWADPVRGSG-----------
Rv1931c|M.tuberculosis_H37Rv ----TGRRRPRHRDCPDGCPLARPVSAPTRWADPVRGSG-----------
MAV_4981|M.avium_104 ALFVCDGDVFTSAGISSGIDLALALVEMDYGTELVRAVARWLVVYLKRAG
TH_2340|M.thermoresistible__bu EMMIYALRWPIDRPHGDEISDTFFRTLAHLVSEALDHEAP----------
Mflv_2899|M.gilvum_PYR-GCK -------------------MRAKDLVDARYAEPITVDDLAAAAGLSRAHF
Mvan_3612|M.vanbaalenii_PYR-1 -----------MLPPARYLMRAKDLVDARYSEPITVDDLAAAAGLSRAHF
MAB_4709c|M.abscessus_ATCC_199 ---------EQRLRDLARLRRVRDRIDREYAQPLNVEALAREAQMSAGHL
MSMEG_0213|M.smegmatis_MC2_155 ---------EQRLRDLKLLRRVRDRIDREYAQPLNVEALARGVNMSAGHL
MMAR_1063|M.marinum_M ---------DLRRSGEPLLAEVFEVIDRRHAEPLSLRDVAAEVGMTPGHL
MUL_0821|M.ulcerans_Agy99 ---------DLRRSGEPLLAEVFEVIDRRHAEPLSLRDVAAEVGMTPGHL
Mb1966c|M.bovis_AF2122/97 ---------VDATRQTDLDPPGAGGHRGRAGGAHRIGELAQRAAMSPRHF
Rv1931c|M.tuberculosis_H37Rv ---------VDATRQTDLDPPGAGGHRGRAGGAHRIGELAQRAAMSPRHF
MAV_4981|M.avium_104 GQSQFSALVEAEPPPRSPLRKVTAAISADPAADHSVKSLSARASLSTRQL
TH_2340|M.thermoresistible__bu ---------LSLPTSDHPIVAAAMAYTKEHLQSVTAEEVSRAVAVSERTL
:: . :: :
Mflv_2899|M.gilvum_PYR-GCK SRMFTRTFGESPRAYLQTRRLERAASLLRHTDRSVADICVMVGLQSVGSF
Mvan_3612|M.vanbaalenii_PYR-1 SRMFSKTFGESPRAYLQTRRLERAASLLRHTDRSVADICVMVGLQSVGSF
MAB_4709c|M.abscessus_ATCC_199 SRQFKSAYGESPYSYLMTRRIERAMMLLRRGDMSVTDVCFAVGCSSLGTF
MSMEG_0213|M.smegmatis_MC2_155 SRQFKAAFGESPYSYLMTRRIERAMALLRRGDLSVTEVCFAVGCSSLGTF
MMAR_1063|M.marinum_M TTIVRRRTGRTVQEWIIERRMAQARELLVATDLAVGEVARRVGISDPGYF
MUL_0821|M.ulcerans_Agy99 TTIVRRRTGRTVQEWIIERRMAQARELLVATDLAVGEVARRVGISDPGYF
Mb1966c|M.bovis_AF2122/97 TRVFSDEVGEAPGRYVERIRTEAARRQLEETHDTVVAIAARCGFGTAETM
Rv1931c|M.tuberculosis_H37Rv TRVFSDEVGEAPGRYVERIRTEAARRQLEETHDTVVAIAARCGFGTAETM
MAV_4981|M.avium_104 TRLFQSELGTTPARYVEMVRIDAARAALDAG-RSVAESARLAGFGSAETL
TH_2340|M.thermoresistible__bu RRQFRQATGISWRTYLLHARMLKAMALLAAPDQSVQETASAVGFDSLSAF
. * : :: * * * :* . * :
Mflv_2899|M.gilvum_PYR-GCK TTSFARAYGLPPAAYRASMPSAAVHARIPSCILQRDTRPKADSRV-KTAH
Mvan_3612|M.vanbaalenii_PYR-1 TTSFARTYGMPPAAYRASMPPAAVHARIPSCILQRDTRPKADTRVDKTAH
MAB_4709c|M.abscessus_ATCC_199 STRFAELVGMSPRTYKER--IVD-HAGIPSCVVKQVTRPIRNREATPAVR
MSMEG_0213|M.smegmatis_MC2_155 STRFTELVGLPPSTYRDR--AAAIQEGLPSCLEKYVTRPVRNREA-PAVR
MMAR_1063|M.marinum_M SRLFSRAHGTSPRQWRAL--------------------PPRH--------
MUL_0821|M.ulcerans_Agy99 SRLFSRAHGTSPRQWRAL--------------------PPRH--------
Mb1966c|M.bovis_AF2122/97 RRSFIRRVGISPDQYRKAFA------------------------------
Rv1931c|M.tuberculosis_H37Rv RRSFIRRVGISPDQYRKAFA------------------------------
MAV_4981|M.avium_104 RRVFVEHLGLSPKAYRDRFRTAARG-------------------------
TH_2340|M.thermoresistible__bu TRAFTQFCGETPSAYRHRISTDP------------HSRPTSADPIG----
* . * .* ::
Mflv_2899|M.gilvum_PYR-GCK GEKTGANASP
Mvan_3612|M.vanbaalenii_PYR-1 GEKTG-----
MAB_4709c|M.abscessus_ATCC_199 G---------
MSMEG_0213|M.smegmatis_MC2_155 LHLA------
MMAR_1063|M.marinum_M ----------
MUL_0821|M.ulcerans_Agy99 ----------
Mb1966c|M.bovis_AF2122/97 ----------
Rv1931c|M.tuberculosis_H37Rv ----------
MAV_4981|M.avium_104 ----------
TH_2340|M.thermoresistible__bu ----------