For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAQAARLIQHRSGVPIYGYRTDPDIPRVSVLRPGPEDLGDRARHIHDFPALWYVPAAGLVYIAAAGQVLE PTQFGSLQGGVAMFFDPAALGGDGGSPWPSWRTHPLLFPFLHGHSGGILRLEVPAARRSTWDGAIAAIEA ELTARQQGYGQAVAAHLTLLLIELARLARDVVGDLRRSGEPLLAEVFEVIDRRHAEPLSLRDVAAEVGMT PGHLTTIVRRRTGRTVQEWIIERRMAQARELLVATDLAVGEVARRVGISDPGYFSRLFSRAHGTSPRQWR ALPPRH
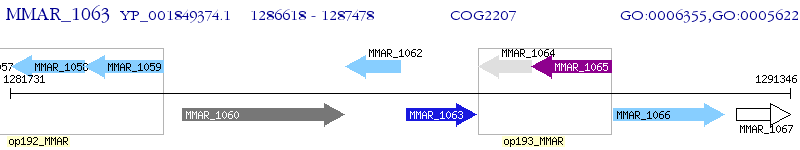
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1063 | - | - | 100% (286) | transcriptional regulatory protein |
| M. marinum M | MMAR_4426 | - | 2e-08 | 28.03% (157) | transcriptional regulatory protein |
| M. marinum M | MMAR_2855 | - | 3e-07 | 28.85% (104) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2899 | - | 1e-12 | 32.99% (97) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2899c | - | 9e-11 | 31.37% (153) | AraC family transcriptional regulator |
| M. avium 104 | MAV_4063 | - | 1e-09 | 31.79% (151) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_3998 | - | 2e-13 | 46.15% (78) | MmsAB operon regulatory protein |
| M. thermoresistible (build 8) | TH_3937 | - | 3e-08 | 33.66% (101) | PUTATIVE putative transcriptional regulator, AraC family |
| M. ulcerans Agy99 | MUL_0821 | - | 1e-164 | 98.60% (286) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5518 | - | 4e-97 | 62.77% (282) | response regulator receiver protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1063|M.marinum_M ---------------------------------MAQAARLIQHRSGVPI-
MUL_0821|M.ulcerans_Agy99 ---------------------------MDYLWRMAQAARLIQHRSGVPI-
Mvan_5518|M.vanbaalenii_PYR-1 ---------------------------------MSETAVFAGRRDGTPV-
Mflv_2899|M.gilvum_PYR-GCK ----------------------------------MRAKDLVDARYAEPI-
MAB_2899c|M.abscessus_ATCC_199 MRSARGPHSGEPGRSGEPHSGRLGRSGEPHSGRLGRSGEPHSGRLGRSG-
MSMEG_3998|M.smegmatis_MC2_155 ---------------------------MDESTAAWIADGFVGQRMLAVP-
TH_3937|M.thermoresistible__bu --------VSDDTMVATAERQGPITRSRTELTDPSEVGDFLEAVYGVRVS
MAV_4063|M.avium_104 -----MGLTTMTVRDDGAPVGAPAEAARSDYGPVDPVLAVNGERRARPVA
MMAR_1063|M.marinum_M YGY-RTDPDIPRVSVLRPGPEDLGDRARHIHDFPALWYVPAAGLVYIAAA
MUL_0821|M.ulcerans_Agy99 YGY-RTDPDIPRVSVRRPGPEDLGDRARHIHDFPALWYVPAAGLVYIAAA
Mvan_5518|M.vanbaalenii_PYR-1 YAYPRRRPQAP-VSVLELGPDAPPVRTTHIHEFPVLVYQPATGSVDVVAA
Mflv_2899|M.gilvum_PYR-GCK TVDDLAAA------------------------------------------
MAB_2899c|M.abscessus_ATCC_199 EPHSVVVLALPGTIGFDLATAVEVFRRVYLADGSRPYLVRVCGTEPVVSA
MSMEG_3998|M.smegmatis_MC2_155 RPVVDHALGAPVTRRLLVTDAGFFPRATRHGRSRSRGATEHVVMVCTAGA
TH_3937|M.thermoresistible__bu RVRPPRSHDGPLLTHVRADAGSFIVDRLHIAGELHTTADPIHKVVTLWPT
MAV_4063|M.avium_104 VEYKQFRGADAQSARRFFATAYDPGWRIAGVTNHCVVSHRRSDTGSMTVD
MMAR_1063|M.marinum_M GQVLEPTQFGSLQGGVAMFFDPAALGG-----------------------
MUL_0821|M.ulcerans_Agy99 GQVLEPTQFGSLQAGVAMFFDPAALGG-----------------------
Mvan_5518|M.vanbaalenii_PYR-1 GLPVDPGRGGDRTGAVAVFFDPAALGE-----------------------
Mflv_2899|M.gilvum_PYR-GCK ------------AGLSRAHFSRMFTRT-----------------------
MAB_2899c|M.abscessus_ATCC_199 GPFGIATDLGLEAVSEAETIVVPARDNPTAPLPEGVLDALRGAYERGTRI
MSMEG_3998|M.smegmatis_MC2_155 GWCRTEEGAFDVGVGDAVLLPKGRAHMYG---------------------
TH_3937|M.thermoresistible__bu SGRVVGRCAGLEGRAGAGEIILSGQPDLVR--------------------
MAV_4063|M.avium_104 EVAIQGRMTLEIPAGDTVVVIQPRAGALNVAGGPLATPDFPLLVAHG---
.
MMAR_1063|M.marinum_M ----------------DGGSPWPSWRTHPLLFPFLHGHSGGILRLEVPAA
MUL_0821|M.ulcerans_Agy99 ----------------DGGSPWPSWRTHPLLFPFLHGHSGGILRLEVPAA
Mvan_5518|M.vanbaalenii_PYR-1 ----------------GARSPWPTWRAHPLLSPFLHGHHDGILRLEVPAA
Mflv_2899|M.gilvum_PYR-GCK ----------------FGESPRAYLQTR---------------RLERAAS
MAB_2899c|M.abscessus_ATCC_199 ASICAGAFTLAEAGILDGKRATTHWLAADLFRERYPAVHLDADVLYVDEG
MSMEG_3998|M.smegmatis_MC2_155 ---------------AAESDPWTLWWMHVTGSDSAELIAAARVSPVSPVV
TH_3937|M.thermoresistible__bu ---QTHCDDLAATVILLDPALVAHVASGSAGDDAAAAIRFASLRPAVGAT
MAV_4063|M.avium_104 MACVLHCHGARFDVVTIAAEVLQKVAAEWHTALSQQTQFLNWRPRSRAAV
MMAR_1063|M.marinum_M RRSTWDGAIAAIEAELTARQQGYGQAVAAHLTLLLIELARL---ARDVVG
MUL_0821|M.ulcerans_Agy99 RRSTWDGAIAAIEAELTTRQQGYGQAVTAHLTLLLIELARL---ARDVVG
Mvan_5518|M.vanbaalenii_PYR-1 RRPFWDAGVAALQAELTDRRDGYAQAALAHLTVLLIELARL---SDDVVG
Mflv_2899|M.gilvum_PYR-GCK LLRHTDRSVADICVMVGLQS----------------------------VG
MAB_2899c|M.abscessus_ATCC_199 QVLTSAGAAAGLDLCLHIVARDHGAAVAAEAARVAVAPLHRDGGQAQYVI
MSMEG_3998|M.smegmatis_MC2_155 HLVDPAPATSLISQVIDMLADASTTAGLIGAAGAAWHALTV---IAATGR
TH_3937|M.thermoresistible__bu RLWQDTVRYVRDHVLVDDTRATPLVVGQAARLLAAVTASAFPNSAAEPAG
MAV_4063|M.avium_104 RAWHRALDYAMLTLASPDTARQPLIAAGMAGLLAGALLECYP-SNLTEQD
MMAR_1063|M.marinum_M DLRRSGEP-LLAEVFEVIDRRHAEPLSLRDVAAEVGMTPGHLTTIVRRRT
MUL_0821|M.ulcerans_Agy99 DLRRSGEP-LLAEVFEVIDRRHAEPLSLRDVAAEVGMTPGHLTTIVRRRT
Mvan_5518|M.vanbaalenii_PYR-1 DLRRSNEP-LLAQVFDVIERRLGEPLTLRDVARECGLTAGYLTTVVRRRT
Mflv_2899|M.gilvum_PYR-GCK SFTTS------------FARAYGLPPAAYRASMPSAAVHARIPSCILQRD
MAB_2899c|M.abscessus_ATCC_199 RNRRRAEV-SLGEILAWIEHNAHRELSLEDLATEASTSVRSLNRRFRAET
MSMEG_3998|M.smegmatis_MC2_155 QRSTGEAD-PVDRALEHLRSTSPRRTDVAALAAIVGLSTSHLNALFRQRT
TH_3937|M.thermoresistible__bu QTVNGQRPPLLRRALDYIDANAAADISLADIAAAVHVTPRALQYMFRRHL
MAV_4063|M.avium_104 PVSDLALPETLKEAVSFIHRHAAEDIGINDVAAAVHLTPRAVQYLFRRQL
: : : . .
MMAR_1063|M.marinum_M GRTVQEWIIERRMAQARELLVATD---LAVGEVARRVGISDPGYFSRLFS
MUL_0821|M.ulcerans_Agy99 GRTVQEWIIERRMAQARELLVATD---LAVGEVARRVGISDPGYFSRLFS
Mvan_5518|M.vanbaalenii_PYR-1 GRTVGSWIAERRMAEARALLLATE---LTVGEVGRRVGMTDPGYFTRQFR
Mflv_2899|M.gilvum_PYR-GCK TRPKADSRVKTAHGEKTGANASP---------------------------
MAB_2899c|M.abscessus_ATCC_199 GQTPMQWLTGVRVRHAQQLLERSA---DSVEWIGREVGFGSPANFREQFR
MSMEG_3998|M.smegmatis_MC2_155 GIGPLEFQQQLRMSRARELLDTTT---LTIAAVARAAGFNDPLYFSRQFA
TH_3937|M.thermoresistible__bu QTTPLQYLRRIRLHRAHHDLLAQTPASDTVGAIAARWGFAHAGRFAMQYR
MAV_4063|M.avium_104 DTTPTEYMRRVRLSRAHQELVAATTASSTVTEIAQRWGFAHTGRFAVLYR
. .
MMAR_1063|M.marinum_M RAHGTSPRQWRALPPRH--
MUL_0821|M.ulcerans_Agy99 RAHGTSPRQWRALPPRH--
Mvan_5518|M.vanbaalenii_PYR-1 SCHGESPRTWRARHG----
Mflv_2899|M.gilvum_PYR-GCK -------------------
MAB_2899c|M.abscessus_ATCC_199 RLTGVSPLAYRNTFRAQSA
MSMEG_3998|M.smegmatis_MC2_155 KVHGQSPSAYRQRHQN---
TH_3937|M.thermoresistible__bu QTYGRSPHQTLRG------
MAV_4063|M.avium_104 QTYGQSPHTTLRQQTAV--