For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PGEEQRLRDLKLLRRVRDRIDREYAQPLNVEALARGVNMSAGHLSRQFKAAFGESPYSYLMTRRIERAMA LLRRGDLSVTEVCFAVGCSSLGTFSTRFTELVGLPPSTYRDRAAAIQEGLPSCLEKYVTRPVRNREAPAV RLHLA*
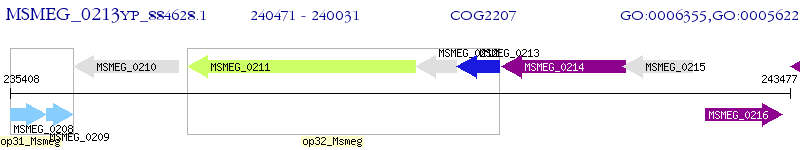
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0213 | - | - | 100% (146) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6356 | - | 1e-10 | 32.11% (109) | AraC family transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_6546 | - | 4e-10 | 38.04% (92) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2899 | - | 7e-27 | 46.22% (119) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4709c | - | 4e-59 | 80.14% (141) | AraC family transcriptional regulator |
| M. marinum M | MMAR_1063 | - | 7e-11 | 36.36% (99) | transcriptional regulatory protein |
| M. avium 104 | MAV_4981 | - | 2e-08 | 33.64% (107) | transcriptional regulator, AraC family protein |
| M. thermoresistible (build 8) | TH_2340 | - | 3e-13 | 39.60% (101) | transcriptional regulatory protein |
| M. ulcerans Agy99 | MUL_0821 | - | 5e-11 | 36.36% (99) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3008 | - | 7e-64 | 79.87% (149) | helix-turn-helix domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0213|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3008|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1063|M.marinum_M -------------------------MAQAARLIQHRSGVPIYGYRTDPDI
MUL_0821|M.ulcerans_Agy99 -------------------MDYLWRMAQAARLIQHRSGVPIYGYRTDPDI
MAV_4981|M.avium_104 MADSSATPRVVVIVVFDDVTMLDVAGAGEVFAEANRFGADYLLKIASVDG
TH_2340|M.thermoresistible__bu ---------------------------------VTVSALSARNLASSAKP
MSMEG_0213|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3008|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1063|M.marinum_M PRVSVLRPGPEDLGDRARHIHDFPALWYVPAAGLVYIAAAGQVLEPTQFG
MUL_0821|M.ulcerans_Agy99 PRVSVRRPGPEDLGDRARHIHDFPALWYVPAAGLVYIAAAGQVLEPTQFG
MAV_4981|M.avium_104 RDVTTSIGTRLGVTDAVSSIESADTVLVAGSDNLPRRPIDPELVEAVKSV
TH_2340|M.thermoresistible__bu PQIDLRRGGRALGGSYLYEGDKLQTGWHSHDVHQIEYAIGGVVEVQTRTG
MSMEG_0213|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3008|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1063|M.marinum_M SLQGG------VAMFFDPAALGGDGGSPWPSWRTHPLLFPFLHGHSGGIL
MUL_0821|M.ulcerans_Agy99 SLQAG------VAMFFDPAALGGDGGSPWPSWRTHPLLFPFLHGHSGGIL
MAV_4981|M.avium_104 ADRTRRLASICTGSFILGQAGLLDGRRATTHWHDVRLFARAFPEITVEPD
TH_2340|M.thermoresistible__bu HYLLPPQQAAWLPVGLEHSATMNPDVKTVAVMFDPQLVPDAGNRARILAV
MSMEG_0213|M.smegmatis_MC2_155 -----------------------------------------------MPG
Mvan_3008|M.vanbaalenii_PYR-1 -----------------------------------------------MPG
MAB_4709c|M.abscessus_ATCC_199 ------------------------------------MLFGTNLGTVTSAG
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1063|M.marinum_M RLEVPAARRSTWDGAIAAIEAELTARQQGYGQAVAAHLTLLLIELARLAR
MUL_0821|M.ulcerans_Agy99 RLEVPAARRSTWDGAIAAIEAELTTRQQGYGQAVTAHLTLLLIELARLAR
MAV_4981|M.avium_104 ALFVCDGDVFTSAGISSGIDLALALVEMDYGTELVRAVARWLVVYLKRAG
TH_2340|M.thermoresistible__bu SPLIREMMIYALR----------WPIDRPHGDEISDTFFRTLAHLVSEAL
MSMEG_0213|M.smegmatis_MC2_155 E---EQRLRDL-----KLLRRVRDRIDREYAQPLNVEALARGVNMSAGHL
Mvan_3008|M.vanbaalenii_PYR-1 ESVSAQRLRDL-----ALLRRVKDRIDREYAQPLNVEALARGVNMSAGHL
MAB_4709c|M.abscessus_ATCC_199 E---EQRLRDL-----ARLRRVRDRIDREYAQPLNVEALAREAQMSAGHL
Mflv_2899|M.gilvum_PYR-GCK -------------------MRAKDLVDARYAEPITVDDLAAAAGLSRAHF
MMAR_1063|M.marinum_M DVVGDLRRSGE-----PLLAEVFEVIDRRHAEPLSLRDVAAEVGMTPGHL
MUL_0821|M.ulcerans_Agy99 DVVGDLRRSGE-----PLLAEVFEVIDRRHAEPLSLRDVAAEVGMTPGHL
MAV_4981|M.avium_104 GQSQFSALVEAEPPPRSPLRKVTAAISADPAADHSVKSLSARASLSTRQL
TH_2340|M.thermoresistible__bu DHEAPLSLPTS----DHPIVAAAMAYTKEHLQSVTAEEVSRAVAVSERTL
. . :: . :: :
MSMEG_0213|M.smegmatis_MC2_155 SRQFKAAFGESPYSYLMTRRIERAMALLRRGDLSVTEVCFAVGCSSLGTF
Mvan_3008|M.vanbaalenii_PYR-1 SRQFKSAYGESPYTYLMTRRIERAMALLRRGDMSVTEVCFEVGCASLGTF
MAB_4709c|M.abscessus_ATCC_199 SRQFKSAYGESPYSYLMTRRIERAMMLLRRGDMSVTDVCFAVGCSSLGTF
Mflv_2899|M.gilvum_PYR-GCK SRMFTRTFGESPRAYLQTRRLERAASLLRHTDRSVADICVMVGLQSVGSF
MMAR_1063|M.marinum_M TTIVRRRTGRTVQEWIIERRMAQARELLVATDLAVGEVARRVGISDPGYF
MUL_0821|M.ulcerans_Agy99 TTIVRRRTGRTVQEWIIERRMAQARELLVATDLAVGEVARRVGISDPGYF
MAV_4981|M.avium_104 TRLFQSELGTTPARYVEMVRIDAARAALDAG-RSVAESARLAGFGSAETL
TH_2340|M.thermoresistible__bu RRQFRQATGISWRTYLLHARMLKAMALLAAPDQSVQETASAVGFDSLSAF
. * : :: *: * * :* : . .* . :
MSMEG_0213|M.smegmatis_MC2_155 STRFTELVGLPPSTYRDRA--AAIQEGLPSCLEKYVTRPVRNREAPAVRL
Mvan_3008|M.vanbaalenii_PYR-1 STRFTELVGVPPSVYRRDA--AEVAPGIPSCVAKQVTRPIRNREAPVSAL
MAB_4709c|M.abscessus_ATCC_199 STRFAELVGMSPRTYKERI--VDHA-GIPSCVVKQVTRPIRNREATPAVR
Mflv_2899|M.gilvum_PYR-GCK TTSFARAYGLPPAAYRASMPSAAVHARIPSCILQRDTRPKADSRVKTAHG
MMAR_1063|M.marinum_M SRLFSRAHGTSPRQWRALP---------PRH-------------------
MUL_0821|M.ulcerans_Agy99 SRLFSRAHGTSPRQWRALP---------PRH-------------------
MAV_4981|M.avium_104 RRVFVEHLGLSPKAYRDRF--------------RTAARG-----------
TH_2340|M.thermoresistible__bu TRAFTQFCGETPSAYRHRIS------------TDPHSRPTSADPIG----
* . * .* ::
MSMEG_0213|M.smegmatis_MC2_155 HLA------
Mvan_3008|M.vanbaalenii_PYR-1 PLA------
MAB_4709c|M.abscessus_ATCC_199 G--------
Mflv_2899|M.gilvum_PYR-GCK EKTGANASP
MMAR_1063|M.marinum_M ---------
MUL_0821|M.ulcerans_Agy99 ---------
MAV_4981|M.avium_104 ---------
TH_2340|M.thermoresistible__bu ---------