For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LDYLWRMAQAARLIQHRSGVPIYGYRTDPDIPRVSVRRPGPEDLGDRARHIHDFPALWYVPAAGLVYIAA AGQVLEPTQFGSLQAGVAMFFDPAALGGDGGSPWPSWRTHPLLFPFLHGHSGGILRLEVPAARRSTWDGA IAAIEAELTTRQQGYGQAVTAHLTLLLIELARLARDVVGDLRRSGEPLLAEVFEVIDRRHAEPLSLRDVA AEVGMTPGHLTTIVRRRTGRTVQEWIIERRMAQARELLVATDLAVGEVARRVGISDPGYFSRLFSRAHGT SPRQWRALPPRH
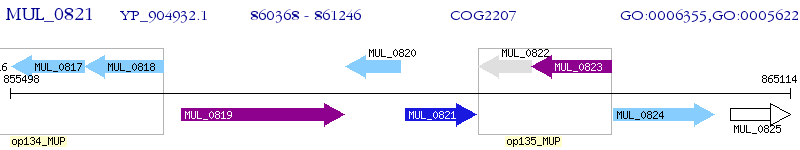
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0821 | - | - | 100% (292) | transcriptional regulatory protein |
| M. ulcerans Agy99 | MUL_2902 | - | 7e-07 | 28.85% (104) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2899 | - | 1e-12 | 32.99% (97) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4709c | - | 2e-10 | 34.69% (98) | AraC family transcriptional regulator |
| M. marinum M | MMAR_1063 | - | 1e-164 | 98.60% (286) | transcriptional regulatory protein |
| M. avium 104 | MAV_4063 | - | 7e-09 | 35.05% (97) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_3998 | - | 2e-13 | 46.15% (78) | MmsAB operon regulatory protein |
| M. thermoresistible (build 8) | TH_3937 | - | 4e-08 | 33.66% (101) | PUTATIVE putative transcriptional regulator, AraC family |
| M. vanbaalenii PYR-1 | Mvan_5518 | - | 9e-96 | 62.06% (282) | response regulator receiver protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0821|M.ulcerans_Agy99 -----------------------MDYLWRMAQAARLIQHRSGVPIYGY-R
MMAR_1063|M.marinum_M -----------------------------MAQAARLIQHRSGVPIYGY-R
Mvan_5518|M.vanbaalenii_PYR-1 -----------------------------MSETAVFAGRRDGTPVYAYPR
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3998|M.smegmatis_MC2_155 --------------------MDESTAAWIADGFVGQRMLAVPRPVVDHAL
TH_3937|M.thermoresistible__bu ----------------VSDDTMVATAERQGPITRSRTELTDPSEVGDFLE
MAV_4063|M.avium_104 MGLTTMTVRDDGAPVGAPAEAARSDYGPVDPVLAVNGERRARPVAVEYKQ
MUL_0821|M.ulcerans_Agy99 TDPDIPRVSVRRPGPED-----------------LGDRARHIHDFPALWY
MMAR_1063|M.marinum_M TDPDIPRVSVLRPGPED-----------------LGDRARHIHDFPALWY
Mvan_5518|M.vanbaalenii_PYR-1 RRPQAP-VSVLELGPDA-----------------PPVRTTHIHEFPVLVY
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3998|M.smegmatis_MC2_155 GAPVTRRLLVTDAGFFP-----------------RATRHGRSRSRGATEH
TH_3937|M.thermoresistible__bu AVYGVRVSRVRPPRSHDGPLLTHVRADAGSFIVDRLHIAGELHTTADPIH
MAV_4063|M.avium_104 FRGADAQSARRFFATAYDPGWRIAGVTNHCVVSHRRSDTGSMTVDEVAIQ
MUL_0821|M.ulcerans_Agy99 VPAAGLVYIAAAGQVLEPTQFGSLQA-------GVAMFFDPAALGGDGGS
MMAR_1063|M.marinum_M VPAAGLVYIAAAGQVLEPTQFGSLQG-------GVAMFFDPAALGGDGGS
Mvan_5518|M.vanbaalenii_PYR-1 QPATGSVDVVAAGLPVDPGRGGDRTG-------AVAVFFDPAALGEGARS
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3998|M.smegmatis_MC2_155 VVMVCTAGAGWCRTEEGAFDVGVGDA-------VLLPKGRAHMYGAAESD
TH_3937|M.thermoresistible__bu KVVTLWPTSGRVVGRCAGLEGRAGAG-------EIILSGQPDLVRQTHCD
MAV_4063|M.avium_104 GRMTLEIPAGDTVVVIQPRAGALNVAGGPLATPDFPLLVAHGMACVLHCH
MUL_0821|M.ulcerans_Agy99 PWPSWRTHPLLFPFLHGHSGGILRLEVPAARRSTWDGAI-----------
MMAR_1063|M.marinum_M PWPSWRTHPLLFPFLHGHSGGILRLEVPAARRSTWDGAI-----------
Mvan_5518|M.vanbaalenii_PYR-1 PWPTWRAHPLLSPFLHGHHDGILRLEVPAARRPFWDAGV-----------
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_3998|M.smegmatis_MC2_155 PWTLWWMHVTGSDSAELIAAARVSPVSPVVHLVDPAPAT-----------
TH_3937|M.thermoresistible__bu DLAATVILLDPALVAHVASGSAGDDAAAAIRFASLRPAVGATRLWQDTVR
MAV_4063|M.avium_104 GARFDVVTIAAEVLQKVAAEWHTALSQQTQFLNWRPRSRAAVRAWHRALD
MUL_0821|M.ulcerans_Agy99 -----AAIEAELTTRQQGYGQAVTAHLTLLLIELARLARDVVGDLRRSGE
MMAR_1063|M.marinum_M -----AAIEAELTARQQGYGQAVAAHLTLLLIELARLARDVVGDLRRSGE
Mvan_5518|M.vanbaalenii_PYR-1 -----AALQAELTDRRDGYAQAALAHLTVLLIELARLSDDVVGDLRRSNE
Mflv_2899|M.gilvum_PYR-GCK --------------------------------------------------
MAB_4709c|M.abscessus_ATCC_199 ----------------------------MLFGTNLGTVTSAGEEQRLRDL
MSMEG_3998|M.smegmatis_MC2_155 -----SLISQVIDMLADASTTAGLIGAAGAAWHALTVIAATGRQRSTGEA
TH_3937|M.thermoresistible__bu YVRDHVLVDDTRATPLVVGQAARLLAAVTASAFPNSAAEPAGQTVNGQRP
MAV_4063|M.avium_104 YAMLTLASPDTARQPLIAAGMAGLLAGALLECYPSNLTEQD-PVSDLALP
MUL_0821|M.ulcerans_Agy99 PLLAEVFEVIDRRHAEPLSLRDVAAEVGMTPGHLTTIVRRRTGRTVQEWI
MMAR_1063|M.marinum_M PLLAEVFEVIDRRHAEPLSLRDVAAEVGMTPGHLTTIVRRRTGRTVQEWI
Mvan_5518|M.vanbaalenii_PYR-1 PLLAQVFDVIERRLGEPLTLRDVARECGLTAGYLTTVVRRRTGRTVGSWI
Mflv_2899|M.gilvum_PYR-GCK ---MRAKDLVDARYAEPITVDDLAAAAGLSRAHFSRMFTRTFGESPRAYL
MAB_4709c|M.abscessus_ATCC_199 ARLRRVRDRIDREYAQPLNVEALAREAQMSAGHLSRQFKSAYGESPYSYL
MSMEG_3998|M.smegmatis_MC2_155 DPVDRALEHLRSTSPRRTDVAALAAIVGLSTSHLNALFRQRTGIGPLEFQ
TH_3937|M.thermoresistible__bu PLLRRALDYIDANAAADISLADIAAAVHVTPRALQYMFRRHLQTTPLQYL
MAV_4063|M.avium_104 ETLKEAVSFIHRHAAEDIGINDVAAAVHLTPRAVQYLFRRQLDTTPTEYM
.. . : : :* :: . . :
MUL_0821|M.ulcerans_Agy99 IERRMAQARELLVATD---LAVGEVARRVGISDPGYFSRLFSRAHGTSPR
MMAR_1063|M.marinum_M IERRMAQARELLVATD---LAVGEVARRVGISDPGYFSRLFSRAHGTSPR
Mvan_5518|M.vanbaalenii_PYR-1 AERRMAEARALLLATE---LTVGEVGRRVGMTDPGYFTRQFRSCHGESPR
Mflv_2899|M.gilvum_PYR-GCK QTRRLERAASLLRHTD---RSVADICVMVGLQSVGSFTTSFARAYGLPPA
MAB_4709c|M.abscessus_ATCC_199 MTRRIERAMMLLRRGD---MSVTDVCFAVGCSSLGTFSTRFAELVGMSPR
MSMEG_3998|M.smegmatis_MC2_155 QQLRMSRARELLDTTT---LTIAAVARAAGFNDPLYFSRQFAKVHGQSPS
TH_3937|M.thermoresistible__bu RRIRLHRAHHDLLAQTPASDTVGAIAARWGFAHAGRFAMQYRQTYGRSPH
MAV_4063|M.avium_104 RRVRLSRAHQELVAATTASSTVTEIAQRWGFAHTGRFAVLYRQTYGQSPH
*: .* * :: : * *: : * .*
MUL_0821|M.ulcerans_Agy99 QWRALPPRH-------------------------------------
MMAR_1063|M.marinum_M QWRALPPRH-------------------------------------
Mvan_5518|M.vanbaalenii_PYR-1 TWRARHG---------------------------------------
Mflv_2899|M.gilvum_PYR-GCK AYRASMPSAAVHARIPSCILQRDTRPKADSRVKTAHGEKTGANASP
MAB_4709c|M.abscessus_ATCC_199 TYKERIVD---HAGIPSCVVKQVTRPIRNR-------EATPAVRG-
MSMEG_3998|M.smegmatis_MC2_155 AYRQRHQN--------------------------------------
TH_3937|M.thermoresistible__bu QTLRG-----------------------------------------
MAV_4063|M.avium_104 TTLRQQTAV-------------------------------------