For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLLEFDADQRLWQETVRDAVSKQCPASLIRGIAENGVDPAPLWKSYLDAGWTELTDPENAVELAIVLEEL GRATDPTPFLATLTQFAPLVGDRFDSGGSGTAVYSGISAIRDAEGWVLTGTDRFVLDGDRADRLAVVTPA GVFVVEAGAVTTRRVDVFDPVLHVAEVSFPGVHVAESDRLAVDTERAHHVALMGMAITTVGACQRILDLV LEHAKSRQQFGVAIGTFQAVQHKATDMYVAIERARALSYFAALTIAADDPRRRLAAAMAKASAGECQALV FKHGIQLFGAMGFTWENDLQFALKRAKAGELLLGGAAEHRAIIAEEYGRDL
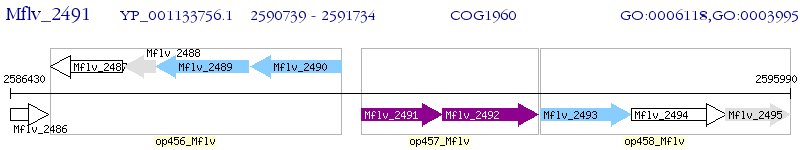
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2491 | - | - | 100% (331) | acyl-CoA dehydrogenase domain-containing protein |
| M. gilvum PYR-GCK | Mflv_0345 | - | 3e-34 | 32.51% (366) | acyl-CoA dehydrogenase domain-containing protein |
| M. gilvum PYR-GCK | Mflv_1478 | - | 7e-30 | 30.63% (333) | acyl-CoA dehydrogenase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1968c | fadE18 | 2e-29 | 33.56% (298) | acyl-CoA dehydrogenase FADE18 |
| M. tuberculosis H37Rv | Rv1933c | fadE18 | 3e-29 | 33.56% (298) | acyl-CoA dehydrogenase FADE18 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 7e-13 | 30.40% (125) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0593c | - | 1e-29 | 31.25% (320) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_4719 | - | 1e-142 | 75.84% (327) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_0934 | - | 1e-149 | 78.90% (327) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4806 | - | 1e-149 | 78.29% (327) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3599 | - | 1e-150 | 78.29% (327) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_0319 | - | 1e-140 | 75.23% (327) | acyl-CoA dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4163 | - | 1e-166 | 88.52% (331) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1968c|M.bovis_AF2122/97 --------------MDFRYSTEQDDFRASLRGFLGR---GAPVREMAAAD
Rv1933c|M.tuberculosis_H37Rv --------------MDFRYSTEQDDFRASLRGFLGR---GAPVREMAAAD
MAB_0593c|M.abscessus_ATCC_199 --------------MSPSGNTERQMLRDTVRSLVTKRADSAAVRTAFESE
Mflv_2491|M.gilvum_PYR-GCK --------------MLLEFDADQRLWQETVRDAVSKQCPASLIRGIAENG
Mvan_4163|M.vanbaalenii_PYR-1 --------------MLLEFDADQKLWQETVRDAVSKQCPASLIRDIAENG
MSMEG_4806|M.smegmatis_MC2_155 --------------MLLEFDADQRLWQETVRDAVTKQCPATLVREVAEQG
TH_3599|M.thermoresistible__bu --------------MLLEFDADQRLWQETVRDAVAKQCPASLIRSIVEDG
MMAR_4719|M.marinum_M --------------MLLEFDADQRLWQATVRDVVTKQCPPSLVRSVAENG
MUL_0319|M.ulcerans_Agy99 MLFSPVSARREAVRMLLEFDADQRLWQATVRDVVTKQCPPSLVRSVAENG
MAV_0934|M.avium_104 --------------MLLEFDADQRLWQSTVRDAVTKQCPPSLVRSVAEEG
MLBr_00737|M.leprae_Br4923 ----MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRAR
:: : ::* :
Mb1968c|M.bovis_AF2122/97 -GSDRRLWQRLCTELELPALHVPPEHGGLGATLVETAIAFAELGRALTPI
Rv1933c|M.tuberculosis_H37Rv -GSDRRLWQRLCTELELPALHVPPEHGGLGATLVETAIAFAELGRALTPI
MAB_0593c|M.abscessus_ATCC_199 RGYDENLWAVLCEQVGAAALLIPEESGGAGGELADAAVVVEELGRGLVPS
Mflv_2491|M.gilvum_PYR-GCK --VDPAPLWKSYLDAGWTELTDPEN-------AVELAIVLEELGRATDPT
Mvan_4163|M.vanbaalenii_PYR-1 --ADATPLWNSYLDAGWTELTDPEN-------AVELAIVLEELGRATDPT
MSMEG_4806|M.smegmatis_MC2_155 --VDPAPLWKNYIEQGWTELTEPEN-------AVELAIVLEELGRATDLT
TH_3599|M.thermoresistible__bu --VDPGPLWKSFVDQGWTELTDPEN-------AVELAIVVEELGRATDPT
MMAR_4719|M.marinum_M --ADSDPLWKVYVDLGWTELNDPAS-------AVELAIVLEELGHATDPT
MUL_0319|M.ulcerans_Agy99 --ADSDPLWKVYVDLGWTELNDPAS-------AVELAIVLEELGHATDPT
MAV_0934|M.avium_104 --VDPSPLWKSYVDQGWTELNDPES-------TVELAIVLEELGHATDPT
MLBr_00737|M.leprae_Br4923 ---FPEEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASA
: * . . .:.. *:.:
Mb1968c|M.bovis_AF2122/97 PFAATVFAI--EAILRMGDDEQRKRLLAGLLTGARIGTIAV---SGHDVA
Rv1933c|M.tuberculosis_H37Rv PFAATVFAI--EAILRMGDDEQRKRLLAGLLTGARIGTIAV---SGHDVA
MAB_0593c|M.abscessus_ATCC_199 PLMGTLWAE--LALLEAGYEG---ELLEALAAGESIGAVAF---DPG---
Mflv_2491|M.gilvum_PYR-GCK PFLATLTQF--APLVGDRFDS----GGSGTAVYSGISAIRD---AEG---
Mvan_4163|M.vanbaalenii_PYR-1 PFLATLTQF--APLVGDRYDS----SVAGTAVYSGVTANRD---AQG---
MSMEG_4806|M.smegmatis_MC2_155 PYLATMTQF--APLAGDRFDA----ASAGAAVYGGVTARRD---ETG---
TH_3599|M.thermoresistible__bu PFLATMTQF--APLVGERFDP----HRAGAAVYGGISAHRD---DTG---
MMAR_4719|M.marinum_M PFLATMSQF--APLAAGCFDP----HQSGTAVYNGITVRRD---AQG---
MUL_0319|M.ulcerans_Agy99 PFLATMSQF--APLAAGCFDP----HQSGTAVYNGITVRRD---AQG---
MAV_0934|M.avium_104 PFLATMSQF--APLVGEHFDP----HQSGTAVYGGVCAYRD---ADG---
MLBr_00737|M.leprae_Br4923 SLIPAVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDA
. :: : : . . :
Mb1968c|M.bovis_AF2122/97 SATTVRAVRRDGRPALTGECTPVLHGHVADLFVVPAVAD---GSIVLHVV
Rv1933c|M.tuberculosis_H37Rv SATTVRAVRRDGRPALTGECTPVLHGHVADLFVVPAVAD---GSIVLHVV
MAB_0593c|M.abscessus_ATCC_199 ---------------------YVVNGDIADVVLT-----------VQGET
Mflv_2491|M.gilvum_PYR-GCK -------------WVLTGTDRFVLDGDRADRLAVVT---------PAGVF
Mvan_4163|M.vanbaalenii_PYR-1 -------------WVLSGTDRFVLDGDRADRLAVVT---------PAGVF
MSMEG_4806|M.smegmatis_MC2_155 -------------WVLSGTGRHVLDGDRAETLAVVT---------EAGVF
TH_3599|M.thermoresistible__bu -------------WILDGTARHVLDADRAEQLAVVT---------SAGVF
MMAR_4719|M.marinum_M -------------WLLDGTARHVLEGDRADRLAVVT---------DAGVF
MUL_0319|M.ulcerans_Agy99 -------------WLLDGTARHVLERDRADRLAVVT---------DAGVF
MAV_0934|M.avium_104 -------------WVLDGTARHVLDGDRVDRLAVVT---------DAGVF
MLBr_00737|M.leprae_Br4923 ASMRTRAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAF
: . .
Mb1968c|M.bovis_AF2122/97 AADAPG----VTVTPL-----PSFDITRPVATLRLAGSPAEPLTAGTPDD
Rv1933c|M.tuberculosis_H37Rv AADAPG----VTVTPL-----PSFDITRPVATLRLAGSPAEPLTAGTPDD
MAB_0593c|M.abscessus_ATCC_199 VCRAGQ----VTALAL-----STLDPTRRLASIAVG--AAEPVGTAP---
Mflv_2491|M.gilvum_PYR-GCK VVEAGA----VTTRRV-----DVFDPVLHVAEVSFPGVHVAESDRLAVD-
Mvan_4163|M.vanbaalenii_PYR-1 CVDPRADSANVTTRRV-----DVFDPVLHVAEVSFAGTQVSDSDRLLVD-
MSMEG_4806|M.smegmatis_MC2_155 LVDAGT----AAIRRS-----PVFDPVLHVAEVSFDGVAVADTDRVPVD-
TH_3599|M.thermoresistible__bu LVDAGV----ATVRRS-----QAFDPVLHITDVSFTGVQVPDTRRLATD-
MMAR_4719|M.marinum_M IVAASQ----VSARRE-----SVFDPVLHVADLSFREVRVPENARVGVD-
MUL_0319|M.ulcerans_Agy99 IVAASQ----VSARRE-----SVFDPVLHVADLSFREVRVPENARVGVD-
MAV_0934|M.avium_104 ITAANQ----VSARRT-----AVFDPVLHVADLSFAEVRVPDSARLSVD-
MLBr_00737|M.leprae_Br4923 IVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKT
. .
Mb1968c|M.bovis_AF2122/97 MERVLDVARVLLAAEMLGGAEACLDLAVQYAGRRTQFDRPIGSFQAVKHA
Rv1933c|M.tuberculosis_H37Rv MERVLDVARVLLAAEMLGGAEACLDLAVQYAGRRTQFDRPIGSFQAVKHA
MAB_0593c|M.abscessus_ATCC_199 --GLIDKAGLLLAAEQVGAAARALDLTVEYTKTRVQFGRAIGSFQALKHR
Mflv_2491|M.gilvum_PYR-GCK TERAHHVALMGMAITTVGACQRILDLVLEHAKSRQQFGVAIGTFQAVQHK
Mvan_4163|M.vanbaalenii_PYR-1 TERARHVALMGMAVTMVGACQRILDLVLEHAKSRQQFGVAIGTFQAVQHK
MSMEG_4806|M.smegmatis_MC2_155 IEKAHHLALTGMAVHTVGACRRVLDLVLEHAKQRQQFGVPIGSLQAVQHK
TH_3599|M.thermoresistible__bu PEKARHVALTGMAMTMVGACQRVLDLALEHVKQRHQFGVPIGSFQAIQHK
MMAR_4719|M.marinum_M AERARHIALTGMAITMVGACQRILDLVLEHVRSRHQFGVPIGSFQAVQHK
MUL_0319|M.ulcerans_Agy99 AERARHIALTGMAITMVGACQRILDLVLEHVRSRHQFGVPIGSFQAVQHK
MAV_0934|M.avium_104 PERAHHVALTGMAITMVGACQRILDLVLEHVKSRQQFGVPIGSFQAVQHK
MLBr_00737|M.leprae_Br4923 ALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFM
. : :. :* . ** .: :. * **. .*.::*:::.
Mb1968c|M.bovis_AF2122/97 CADMMIEIDATRATVMFAAMSAANGDELQ-TVAPLAKAQTAETFVLCAGS
Rv1933c|M.tuberculosis_H37Rv CADMMIEIDATRATVMFAAMSAANGDELQ-TVAPLAKAQTAETFVLCAGS
MAB_0593c|M.abscessus_ATCC_199 MADLYVLVESARATVYQAIHDLTPEG------ATVARIKASEALSAVSGD
Mflv_2491|M.gilvum_PYR-GCK ATDMYVAIERARALSYFAALTIAADDPRRRLAAAMAKASAGECQALVFKH
Mvan_4163|M.vanbaalenii_PYR-1 ATDMFVAIERARALSYFAALTIAADDPRRRLAAAMAKASAGECQSLVFRH
MSMEG_4806|M.smegmatis_MC2_155 AVDMHVAIERARALAYFAALTIAENDPRRRLASRMAKASAGECQAVVFRH
TH_3599|M.thermoresistible__bu AADMHVAIERARALSYFAALTISVDDPRRRLAAAMAKASAGECQSLVFRH
MMAR_4719|M.marinum_M AADMHVAVQRARALAYFAALTISADDPRRRLAAAMAKASAGECQSLVFRH
MUL_0319|M.ulcerans_Agy99 AADMHVAVQRARALAYFAALTFSADDPRRRLAAAMAKASAGECQSLVFLH
MAV_0934|M.avium_104 VADMHVAVQRARALAYFAALTIAADDPRRRLAAAMAKASAGECQSLAFRH
MLBr_00737|M.leprae_Br4923 LADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTTD
.*: : :: :* * : :: :.:
Mb1968c|M.bovis_AF2122/97 ALQIHGAIAFTWEHDLHLYYRRAKTTEALFGSSARNRALLAERAGLVKA-
Rv1933c|M.tuberculosis_H37Rv ALQIHGAIAFTWEHDLHLYYRRAKTTEALFGSSARNRALLAERAGLVKA-
MAB_0593c|M.abscessus_ATCC_199 SIQLHGGIAITWEHDAHLYFKRAHGSALLLGTPRQLLPNLETAAGL----
Mflv_2491|M.gilvum_PYR-GCK GIQLFGAMGFTWENDLQFALKRAKAGELLLGGAAEHRAIIAEEYGRDL--
Mvan_4163|M.vanbaalenii_PYR-1 GVQLFGAMGFTWENDLQFALKRAKAGELMLGGAAEHRAMIAEEYREMV--
MSMEG_4806|M.smegmatis_MC2_155 GLQLFGAMGFTWENDVQFALKRAKAGELLLGGAAEHRAVIAAEYRTTYRG
TH_3599|M.thermoresistible__bu GLQMFGAMGFTWENDLQFALKRAKAGELMLGGAAEHRAVIAEEYRTF---
MMAR_4719|M.marinum_M GLQLFGAMGFTWENDLQFALKRAKAGELMLGGAAQHRVQIAEEYRAADF-
MUL_0319|M.ulcerans_Agy99 GLQLFGAMGFTWGNDLQFALKRAKAGELMLGGAAQHRAQIAEEYRAADF-
MAV_0934|M.avium_104 GLQLHGAMGFTWENDLQFALKRAKAGELMLGGAAEHRARIAEEYRAADF-
MLBr_00737|M.leprae_Br4923 AVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR----
.:*:.*. . * . : *: * :
Mb1968c|M.bovis_AF2122/97 -
Rv1933c|M.tuberculosis_H37Rv -
MAB_0593c|M.abscessus_ATCC_199 -
Mflv_2491|M.gilvum_PYR-GCK -
Mvan_4163|M.vanbaalenii_PYR-1 -
MSMEG_4806|M.smegmatis_MC2_155 L
TH_3599|M.thermoresistible__bu -
MMAR_4719|M.marinum_M -
MUL_0319|M.ulcerans_Agy99 -
MAV_0934|M.avium_104 -
MLBr_00737|M.leprae_Br4923 -