For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SPSGNTERQMLRDTVRSLVTKRADSAAVRTAFESERGYDENLWAVLCEQVGAAALLIPEESGGAGGELAD AAVVVEELGRGLVPSPLMGTLWAELALLEAGYEGELLEALAAGESIGAVAFDPGYVVNGDIADVVLTVQG ETVCRAGQVTALALSTLDPTRRLASIAVGAAEPVGTAPGLIDKAGLLLAAEQVGAAARALDLTVEYTKTR VQFGRAIGSFQALKHRMADLYVLVESARATVYQAIHDLTPEGATVARIKASEALSAVSGDSIQLHGGIAI TWEHDAHLYFKRAHGSALLLGTPRQLLPNLETAAGL*
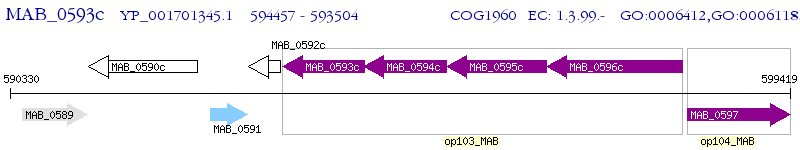
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0593c | - | - | 100% (317) | acyl-CoA dehydrogenase FadE |
| M. abscessus ATCC 19977 | MAB_3123 | - | 7e-39 | 36.99% (346) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0580 | - | 9e-33 | 32.63% (334) | acyl-CoA dehydrogenase FadE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3594 | fadE33 | 1e-104 | 63.52% (307) | acyl-CoA dehydrogenase FADE33 |
| M. gilvum PYR-GCK | Mflv_1477 | - | 1e-101 | 62.54% (307) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3564 | fadE33 | 1e-104 | 63.52% (307) | acyl-CoA dehydrogenase FADE33 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 7e-12 | 32.84% (134) | putative acyl-CoA dehydrogenase |
| M. marinum M | MMAR_5053 | fadE33 | 1e-101 | 62.78% (309) | acyl-CoA dehydrogenase FadE33 |
| M. avium 104 | MAV_0597 | - | 1e-106 | 66.01% (306) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6016 | - | 1e-101 | 62.95% (305) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2066 | fadE33 | 1e-102 | 62.54% (307) | PROBABLE ACYL-CoA DEHYDROGENASE FADE33 |
| M. ulcerans Agy99 | MUL_4129 | fadE33 | 1e-102 | 63.11% (309) | acyl-CoA dehydrogenase FadE33 |
| M. vanbaalenii PYR-1 | Mvan_5289 | - | 1e-100 | 61.44% (306) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1477|M.gilvum_PYR-GCK MS--------------EERELLRDTVAALVEKHASPEAVRTAAASERGYD
Mvan_5289|M.vanbaalenii_PYR-1 MS--------------EERDLLRDTVAALVDKHASPEAVRKAAASERGYD
TH_2066|M.thermoresistible__bu MS--------------EERDLLRDTVATLVDKHATSAAVREAMASERGYD
MSMEG_6016|M.smegmatis_MC2_155 MS--------------EERELLRETVAALVEKHASPEAVRAAMESELGYD
Mb3594|M.bovis_AF2122/97 MTPP------------EERQMLRETVASLVAKHAGPAAVRAAMASDRGYD
Rv3564|M.tuberculosis_H37Rv MTPP------------EERQMLRETVASLVAKHAGPAAVRAAMASDRGYD
MMAR_5053|M.marinum_M MT--------------EEREMLRETVAALVNKHASPAAVRSAMESERGYD
MUL_4129|M.ulcerans_Agy99 MT--------------EEREMLRETVAALVNKHASPAAVRSAMESERGYD
MAV_0597|M.avium_104 MSAIS-----------EERQMLRETVAALVAKHAGPAAVRTAMESERGYD
MAB_0593c|M.abscessus_ATCC_199 MSPSGN----------TERQMLRDTVRSLVTKRADSAAVRTAFESERGYD
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRAR--FP
* *:: ** *: :*. *. . *. : :
Mflv_1477|M.gilvum_PYR-GCK EALWTMLCEQVGAAALVVPEELGGAGGELADAAVVLEELGKRLVPTPLLG
Mvan_5289|M.vanbaalenii_PYR-1 EALWKMLCEQVGAAALVVPEELGGAGGELGDAAVVLEELGKGLVPTPLLG
TH_2066|M.thermoresistible__bu ESLWQLLCEQVGAAALVVPEDFGGAGGELGDAAVVLGELARNLVPTPLFG
MSMEG_6016|M.smegmatis_MC2_155 PNLWRLLCEQVGAAALVIPEEFGGAGGELADAAVVLEELGKALVPTPLLG
Mb3594|M.bovis_AF2122/97 ESLWRLLCEQVGAAALVIPEELGGAGGELADAAIVVQELGRALVPSPLLG
Rv3564|M.tuberculosis_H37Rv ESLWRLLCEQVGAAALVIPEELGGAGGELADAAIVVQELGRALVPSPLLG
MMAR_5053|M.marinum_M ETLWRLLCEQVGAAALVIPEELGGAGGELADAAIVLQELGRALVPSPLLG
MUL_4129|M.ulcerans_Agy99 ETLWRLLCEQVGAAALVIPEELGGAGGELADAAIVLQELGRALVPSPLLG
MAV_0597|M.avium_104 ESLWRLLCEQVGAAALVIPEELGGAGGELADAATVLQELGRALVPSPLLG
MAB_0593c|M.abscessus_ATCC_199 ENLWAVLCEQVGAAALLIPEESGGAGGELADAAVVVEELGRGLVPSPLMG
MLBr_00737|M.leprae_Br4923 EEALAALNAS-GFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIP
* . * *: :**: ** *.: *. *: *:.: ..:.*:
Mflv_1477|M.gilvum_PYR-GCK TT----LAELALLSVDAPDAALLEALAEGTSIGTVVFDP-----------
Mvan_5289|M.vanbaalenii_PYR-1 TT----LAELALLSVAEPDAEALAALAEGAAIGTVVFDP-----------
TH_2066|M.thermoresistible__bu TT----LAELALLASDSPDADALEALAAGASIGAVVFDP-----------
MSMEG_6016|M.smegmatis_MC2_155 TT----LAEIALLSVG--RTEPLEELAEGAKIGTVVFNP-----------
Mb3594|M.bovis_AF2122/97 TT----LAELALLAAAKPDAQALTELAQGSAIGALVLDP-----------
Rv3564|M.tuberculosis_H37Rv TT----LAELALLAAAKPDAQALTELAQGSAIGALVLDP-----------
MMAR_5053|M.marinum_M TT----LAELALLAAAEPDAKTLAGLAEGSSIGALVLDP-----------
MUL_4129|M.ulcerans_Agy99 TT----LAELALLAAAEPDAKTLAGLAEGSSIGALVLDP-----------
MAV_0597|M.avium_104 TT----LAELALLGAAEPDTAALQALAEGSAIGALVLDP-----------
MAB_0593c|M.abscessus_ATCC_199 TL----WAELALLEAG-YEGELLEALAAGESIGAVAFDP-----------
MLBr_00737|M.leprae_Br4923 AVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMR
: : * * ** :.: .:.
Mflv_1477|M.gilvum_PYR-GCK --------EHVVNGNVADVVIGTD---------------GETLSRWTSFT
Mvan_5289|M.vanbaalenii_PYR-1 --------GYVVNGNVADVVIAAD---------------GDALTRWNAFT
TH_2066|M.thermoresistible__bu --------DYVVNGDVAEVIIAAD---------------GERLTRWTGCT
MSMEG_6016|M.smegmatis_MC2_155 --------EFVINGDIADIMIAAD---------------GETLTQWDTFT
Mb3594|M.bovis_AF2122/97 --------DYVVNGDIADIVVAAT---------------SGQLTRWTRFS
Rv3564|M.tuberculosis_H37Rv --------DYVVNGDIADIVVAAT---------------SGQLTRWTRFS
MMAR_5053|M.marinum_M --------DYVLNGDTADVVVAST---------------EGQLSRWTRFS
MUL_4129|M.ulcerans_Agy99 --------DYVLNGDTADVVVAST---------------EGQLSRWTRFS
MAV_0597|M.avium_104 --------DYVVNGDIADVVIGVE---------------DGRLSRWTGVT
MAB_0593c|M.abscessus_ATCC_199 --------GYVVNGDIADVVLTVQ---------------GETVCRAGQVT
MLBr_00737|M.leprae_Br4923 TRAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAFIVHK
::** . : : .
Mflv_1477|M.gilvum_PYR-GCK ATPSVAMDITRPLASLEVTETSPLGTD----PG----------------L
Mvan_5289|M.vanbaalenii_PYR-1 ATPAVAMDITRPLAAVEATETSPLGAD----PG----------------L
TH_2066|M.thermoresistible__bu VEPHQTMDPTRRLARVTPGETTDIGAD----PG----------------L
MSMEG_6016|M.smegmatis_MC2_155 AQPKPTMDMTRRLASVIPGATTTLGDD----QG----------------L
Mb3594|M.bovis_AF2122/97 AQPVATMDPTRRLARLQSEETEPLCPD----PG----------------I
Rv3564|M.tuberculosis_H37Rv AQPVATMDPTRRLARLQSEETEPLCPD----PG----------------I
MMAR_5053|M.marinum_M AHRVATMDPTRRLAQLRAEETAFLGED----PG----------------L
MUL_4129|M.ulcerans_Agy99 AHRVATMDPTRRLAQLRAEETAFLGEN----PG----------------L
MAV_0597|M.avium_104 AQPVTTMDPTRRLARVRAQQTEPIGTD----PG----------------L
MAB_0593c|M.abscessus_ATCC_199 ALALSTLDPTRRLASIAVGAAEPVGTA----PG----------------L
MLBr_00737|M.leprae_Br4923 DDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTALAT
::. .. : : : *
Mflv_1477|M.gilvum_PYR-GCK ADIAAILLAAEQVGAASKCLDLTVQYTKDRVQFGRPIGSFQALKHRMADL
Mvan_5289|M.vanbaalenii_PYR-1 ADTAAILLAAEQIGAATRCLELTVQYTKDRVQFGRPIGSFQALKHRMADL
TH_2066|M.thermoresistible__bu ADTAALLLAAEQIAAAERCLQLTVDYTKERVQFGRPIGSFQALKHRMADL
MSMEG_6016|M.smegmatis_MC2_155 ADTAALLMAAEQIGAASRCLDLTVAYSKDRVQFGRPIGSFQALKHRMADL
Mb3594|M.bovis_AF2122/97 ADTAAILLAAEQIGAAERCLQLTVEYAKSRVQFGRPIGSFQALKHRMADL
Rv3564|M.tuberculosis_H37Rv ADTAAILLAAEQIGAAERCLQLTVEYAKSRVQFGRPIGSFQALKHRMADL
MMAR_5053|M.marinum_M ADAAAILLAAEQIGAAERCLELTVEYAKSRVQFGRPIGSFQALKHRMADL
MUL_4129|M.ulcerans_Agy99 ADAAGILLAAEQIGAAERCLELTVEYAKSRVQFGRPIGSFQALKHRMADL
MAV_0597|M.avium_104 ADTAAVLLAAEQIGAAEQCLARTVEYAKDRVQFGRPIGSFQALKHRMADL
MAB_0593c|M.abscessus_ATCC_199 IDKAGLLLAAEQVGAAARALDLTVEYTKTRVQFGRAIGSFQALKHRMADL
MLBr_00737|M.leprae_Br4923 LDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFMLADM
* : :.*: :. * .* :: *:* * ***..*.:**:::. :**:
Mflv_1477|M.gilvum_PYR-GCK YVAVQSAKAVVDEAVAEPSSTS------AALARLAATEAFSKTAAEAVQM
Mvan_5289|M.vanbaalenii_PYR-1 YVAVQSAKAVVDEAVAEPTATS------AALARLAAGEAFSKVAAEAVQM
TH_2066|M.thermoresistible__bu YVRVQSTRAVVDEAIAEPSATS------AALARFSASETFSAVAAEAVQM
MSMEG_6016|M.smegmatis_MC2_155 YVKVASARAVVHDSIATPSSTS------AALARYFASEALSAVTSEAVQI
Mb3594|M.bovis_AF2122/97 YVTIAAARAVVADACHAPTPTN------AATARLAASEALSTAAAEGIQL
Rv3564|M.tuberculosis_H37Rv YVTIAAARAVVADACHAPTPTN------AATARLAASEALSTAAAEGIQL
MMAR_5053|M.marinum_M YVEVAAARAVVTEACTAPTPTS------AATARLAACEALCAVAAEGIQL
MUL_4129|M.ulcerans_Agy99 YVEVAAARAVVTEACTAPTPTS------AATARLAACEALCAVAAEGIQL
MAV_0597|M.avium_104 YVTVAAAKAVVDEACLDPSPTN------AATARLAATEALNVVAAEGIQL
MAB_0593c|M.abscessus_ATCC_199 YVLVESARATVYQAIHDLTPEG------ATVARIKASEALSAVSGDSIQL
MLBr_00737|M.leprae_Br4923 AMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTTDAVQL
: : ::: * : :: :: * : .: :.:*:
Mflv_1477|M.gilvum_PYR-GCK HGGIAITWESDIQLYFKRAHGSAQLLPPPREQLRRLESEVF-------
Mvan_5289|M.vanbaalenii_PYR-1 HGGIAITWESDIQLYFKRAHGSAQLFGPPREQLRRLEAQVF-------
TH_2066|M.thermoresistible__bu HGGIAITWEHDIQLYFKRAHGSAQLFGPPREHLRRLESEIL-------
MSMEG_6016|M.smegmatis_MC2_155 HGGIAITWEHDIQLYFKRAHGSAQLLGPPREQLRRLEAEVF-------
Mb3594|M.bovis_AF2122/97 HGGIAITWEHDMHLYFKRAHGSAQLLESPREVLRRLESEVWES-P---
Rv3564|M.tuberculosis_H37Rv HGGIAITWEHDMHLYFKRAHGSAQLLESPREVLRRLESEVWES-P---
MMAR_5053|M.marinum_M HGGIAITWEHDMHLYFKRAHGSTHLLASPPELLRRLESEALGAGPSIP
MUL_4129|M.ulcerans_Agy99 HGGIAITWEHDMHLYFKRAHGSTHLLASPPELLRRLESEALGAGPSIP
MAV_0597|M.avium_104 HGGIAITWEHDMHLYFKRAHGSAHLLDTPQQLLTRLEPEVLAT-P---
MAB_0593c|M.abscessus_ATCC_199 HGGIAITWEHDAHLYFKRAHGSALLLGTPRQLLPNLETAAGL------
MLBr_00737|M.leprae_Br4923 FGGAGYTSDFPVERFMRDAK-ITQIYEGTNQIQRVVMSRALLR-----
.** . * : . ::: *: : : . : : .