For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGRVEGKVAFITGAARGQGRAHAVRLAEEGADIIAVDICKQIDSVLIPLSTPEDLAETADLVKNAGGRIH TAEVDVRDYDALKAAVDAGVEEFGRLDIIVANAGIGNGGQTLDKTDEPDWDDMIGVNLSGVWKTVKAGVP HILAGGNGGSIILTSSVGGLKAYPHTGHYVAAKHGVVGLMRTFAVELGAQNIRVNSVHPTNVNTPLFMNE PTMKLFRPDLENPGPDDMKVVGQLMHTLPIGWVEPEDIANAVLFLASDEARFITGVTLPVDGGSCLK*
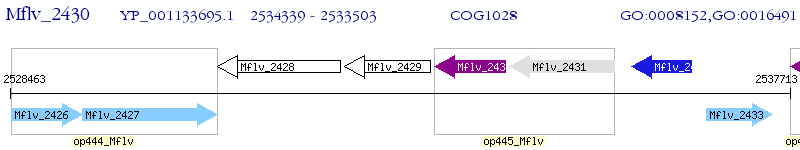
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2430 | - | - | 100% (278) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_1589 | - | e-124 | 77.06% (279) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_2463 | - | e-110 | 65.83% (278) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2771 | - | 1e-72 | 55.11% (274) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. tuberculosis H37Rv | Rv2750 | - | 1e-72 | 55.11% (274) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 2e-23 | 34.65% (202) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0072c | - | 2e-78 | 53.82% (275) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_4754 | - | 1e-125 | 78.85% (279) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_0846 | - | 1e-127 | 79.93% (279) | carveol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6031 | - | 1e-131 | 81.72% (279) | carveol dehydrogenase |
| M. thermoresistible (build 8) | TH_3237 | - | 1e-144 | 89.21% (278) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3392 | fabG | 3e-63 | 48.04% (281) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_4222 | - | 1e-155 | 96.76% (278) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2771|M.bovis_AF2122/97 ---MIDRPLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICEQIAS-
Rv2750|M.tuberculosis_H37Rv ---MIDRPLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICEQIAS-
MUL_3392|M.ulcerans_Agy99 MSRPLDGPLSGRVAFITGAARGQGRAHALRLARDGADVIAVDLCDQIAS-
Mflv_2430|M.gilvum_PYR-GCK ----MAGRVEGKVAFITGAARGQGRAHAVRLAEEGADIIAVDICKQIDS-
Mvan_4222|M.vanbaalenii_PYR-1 ----MAGRVEGKVAFITGAARGQGRAHAVRLAQEGADIIAVDICKQIDS-
TH_3237|M.thermoresistible__bu ----MAGRLEGKVAFITGAARGQGRAHAVRMAQEGADIIAVDICRQIDS-
MAV_0846|M.avium_104 ----MGGRVEGKVAFITGAARGQGRSHAVRLAQEGADIIAVDICAPISSN
MSMEG_6031|M.smegmatis_MC2_155 ----MAGRVEGKVAFITGAARGQGRSHAVRLAEEGADIIAVDVCRRISSN
MMAR_4754|M.marinum_M MTEQLGGRVAGKVAFITGAARGQGRSHAVRLAQEGADIIAVDVCKPIVEN
MAB_0072c|M.abscessus_ATCC_199 ----MTGRLTGKVALVTGAARGIGRAQAVRFAQEGADIIALDICAPVHT-
MLBr_01807|M.leprae_Br4923 ---------MTDAAAKVSAAPGRGK-----PAFVSRSILVTGGNRGIGLA
.* ..** * *: * . .::. . :
Mb2771|M.bovis_AF2122/97 VPYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLSVALQAGLDEFG
Rv2750|M.tuberculosis_H37Rv VPYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLSVALQAGLDEFG
MUL_3392|M.ulcerans_Agy99 VPYPLGTAEELATTVKLVEDTGARIVASQADVRDREALAAALQAGIDELG
Mflv_2430|M.gilvum_PYR-GCK VLIPLSTPEDLAETADLVKNAGGRIHTAEVDVRDYDALKAAVDAGVEEFG
Mvan_4222|M.vanbaalenii_PYR-1 VLIPLSSPEDLAETADLVKNAGGRIHTAEVDVRDFDALKAAVDAGVEEFG
TH_3237|M.thermoresistible__bu VQIPLASPEDLAETADLVKGLNRRIYTAEVDVRDFDALKAAVDTGVEQLG
MAV_0846|M.avium_104 SQIPPSTPDDLAETADLIKGLDRRIVTAEVDVRDYDALKAAVDSGVEQLG
MSMEG_6031|M.smegmatis_MC2_155 EDIPASTPEDLAETVELVKGLNRRIVAEEVDVRDYDALKAVVDSGVEQLG
MMAR_4754|M.marinum_M TTIPASTPEDLAETADLVKGHNRRIFTAEADVRDYDALKAAVDAGVDELG
MAB_0072c|M.abscessus_ATCC_199 TITPPATRADLDKTSELVESAGRRVIPGVVDVRNLRDVQTFTQDAVTEFG
MLBr_01807|M.leprae_Br4923 VARRLVADGHRVAVTHRASVVPDWFFGVECDVTDNDAIGAAFKAVEEHQG
: . . . . . ** : : . . . *
Mb2771|M.bovis_AF2122/97 RLDIVVANAGIAMMQ-----AGDDGWRDVIDVNLTGVFHTVQVAIPTLIE
Rv2750|M.tuberculosis_H37Rv RLDIVVANAGIAMMQ-----AGDDGWRDVIDVNLTGVFHTVQVAIPTLIE
MUL_3392|M.ulcerans_Agy99 QVDIVVANAGIAPMQ-----SGDDGWRDVIDVNLSGAYYTVEVAIPTMIE
Mflv_2430|M.gilvum_PYR-GCK RLDIIVANAGIGNGGQTLDKTDEPDWDDMIGVNLSGVWKTVKAGVPHILA
Mvan_4222|M.vanbaalenii_PYR-1 RLDIIVANAGIGNGGQLLHDTGEPDWDDMIGVNLSGVWKTVKAGVPHILA
TH_3237|M.thermoresistible__bu RLDIIVANAGIGNGGETLDKTSEGDWTDMIDVNLAGVWKTVKAGVPHILA
MAV_0846|M.avium_104 RLDIICANAGIGNGGQTLDKTSEDDWRDMIDVNLSGVWKTVKAGVPHLIS
MSMEG_6031|M.smegmatis_MC2_155 GLDIVVANAGIGNGGATLDKTSEADWDDMIGVNLSGVWKTVKAAVPHLIS
MMAR_4754|M.marinum_M RLDIIVANAGIGNGGATLDKTSEHDWQEMIDVNLSGVWKSVKAAVPHLIA
MAB_0072c|M.abscessus_ATCC_199 GLDIVCATAGITSRRMLLDMP-ESDWQTMLDVNLTGVWHTTRAATPHLIA
MLBr_01807|M.leprae_Br4923 PVEVLVANAGITSDMLIMR-MSEEQFQKVIDVNLTGVFRVAKLASRNMIK
:::: *.*** : : ::.***:*.: .. . ::
Mb2771|M.bovis_AF2122/97 QGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYANHLAPQ
Rv2750|M.tuberculosis_H37Rv QGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYANHLAPQ
MUL_3392|M.ulcerans_Agy99 QGRGGSIVLISSAAGLVGISSADAGAIGYVASKHALVGLMRVYANLLAPH
Mflv_2430|M.gilvum_PYR-GCK GGNGGSIILTSSVGGLKAYPHTG----HYVAAKHGVVGLMRTFAVELGAQ
Mvan_4222|M.vanbaalenii_PYR-1 GGNGGSIILTSSVGGLKAYPHTG----HYVAAKHGVVGLMRTFAVELGAQ
TH_3237|M.thermoresistible__bu GGRGGSIILTSSVGGLKAYPHTG----HYVAAKHGVVGLMRTFAVELGQH
MAV_0846|M.avium_104 QGQGGSVILTSSVGGLKAYPHTG----HYIAAKHGVVGLMRTFAVELGQH
MSMEG_6031|M.smegmatis_MC2_155 GGNGGSIILTSSVGGLKAYPHTG----HYIAAKHGVVGLMRTFAVELGQH
MMAR_4754|M.marinum_M GGNGGSIVLTSSVGGMKAYPHCG----NYVAAKHGVVGLMRSFAVELGQH
MAB_0072c|M.abscessus_ATCC_199 RG-GGAMVLVSSIAGLRGLVGVS----HYVAAKHGVVGLMRSLAIELAPH
MLBr_01807|M.leprae_Br4923 QRFGRYIFMG-SVVGLSGVPGQAN----YAASKSGLIGMARSLARELGSR
* :.: * *: . * *:* .::*: * * *. :
Mb2771|M.bovis_AF2122/97 NIRVNSVHPCGVDTPMINNEFFQQWLT-------TADMDAPHNLGNALPV
Rv2750|M.tuberculosis_H37Rv NIRVNSVHPCGVDTPMINNEFFQQWLT-------TADMDAPHNLGNALPV
MUL_3392|M.ulcerans_Agy99 SIRVNSLHPSGVDPPMINNEFIRHWLAD-----LVAETGSGPGAGNALPV
Mflv_2430|M.gilvum_PYR-GCK NIRVNSVHPTNVNTPLFMNEPTMKLFRPDLENPGPDDMKVVGQLMHTLPI
Mvan_4222|M.vanbaalenii_PYR-1 NVRVNSVHPTNVNTPLFMNEPTMKLFRPDLENPGPEDMKVVGQLMHTLPI
TH_3237|M.thermoresistible__bu NIRVNSVHPTNVNTPLFMNEPTMKLFRPDLENPGPEDLKVVAQMMHTLPI
MAV_0846|M.avium_104 FIRVNSVHPTNVNTPMFMNEGTMRLFRPDLKNPGPDDLEVAAQFMHVLPV
MSMEG_6031|M.smegmatis_MC2_155 SIRVNSVHPTNVNTPLFMNEGTMKLFRPDLENPGPDDMAVVAQMMHVLPV
MMAR_4754|M.marinum_M MIRVNSVHPTHVRTPMLHNEGTFKMFRPDLENPGPDDMAPICQLFHTLPI
MAB_0072c|M.abscessus_ATCC_199 KIRVNTVHPTNVDTPMIQNEFVRTLFRPDLEQPTREDFAAAAKTMNLLDL
MLBr_01807|M.leprae_Br4923 SITANVVAPGFIDTEMTR----------------AMDSKYQEAALQAIPL
: .* : * : . : : : : :
Mb2771|M.bovis_AF2122/97 ELVQP-TDIANAVAWLASEEARYVTGVTLPVDAGFVNKR
Rv2750|M.tuberculosis_H37Rv ELVQP-TDIANAVAWLASEEARYVTGVTLPVDAGFVNKR
MUL_3392|M.ulcerans_Agy99 QILQA-DDIAGALAWLVSDEARYITGVALPVDAGCVNKR
Mflv_2430|M.gilvum_PYR-GCK GWVEP-EDIANAVLFLASDEARFITGVTLPVDGGSCLK-
Mvan_4222|M.vanbaalenii_PYR-1 GWVEP-EDIANAVLFLASDEARFITGVTLPVDGGSCLK-
TH_3237|M.thermoresistible__bu GWVEP-EDIANAVLFLASDEARYITGVTLPVDGGSCLK-
MAV_0846|M.avium_104 GWVEP-VDISNAVLFLASDESRYITGLPVTVDAGSMLK-
MSMEG_6031|M.smegmatis_MC2_155 GWVEP-RDISNAVLFLASDEARYVTGLPMTVDAGSMLK-
MMAR_4754|M.marinum_M PWVEA-EDISNAVLFLASDESRYITGVTLPVDAGGCLK-
MAB_0072c|M.abscessus_ATCC_199 PWVEP-LDIANASLFLASDEARYITSVSLPVDAGSTQR-
MLBr_01807|M.leprae_Br4923 GRIGAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH
: . :*:.* :*.*::: :::. :.**.*