For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSGRVEGKRVLITGAARGMGRSHAVRLAQEGADLILVDICESLPEVEYPLAAVEELADTAELVAQHGRRA VTHVVDVRDEQALSAAVDDGVARLGGLDAAVANAGVLTGGTWDTTTAQQWRTVIDVNLIGTWNTCRAAIP HLLERGGSLINVSSAAGIKGSPLHTPYTASKAGVVGMSKALANELAAMNIRVNTVHPTGVPTGMRPASLH QLLHDTRSDLSPIFQNALPIVMAEAVDISNAVLFLISDEARHVTGLEFKVDAGVTIR
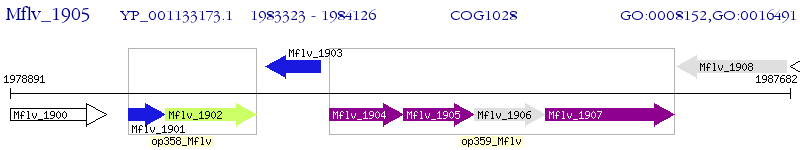
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1905 | - | - | 100% (267) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_2430 | - | 4e-63 | 50.00% (278) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_1589 | - | 1e-61 | 48.75% (279) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2771 | - | 1e-57 | 48.12% (266) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. tuberculosis H37Rv | Rv2750 | - | 1e-57 | 48.12% (266) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01740 | - | 9e-26 | 37.69% (199) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3081 | - | 2e-58 | 45.79% (273) | Short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4754 | - | 1e-63 | 50.90% (279) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_3551 | - | 1e-106 | 71.54% (267) | carveol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5813 | - | 1e-124 | 79.40% (267) | carveol dehydrogenase |
| M. thermoresistible (build 8) | TH_3237 | - | 4e-66 | 51.08% (278) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3392 | fabG | 1e-54 | 44.49% (272) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_3395 | - | 1e-123 | 80.83% (266) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2771|M.bovis_AF2122/97 ----MIDRPLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICE-QIA
Rv2750|M.tuberculosis_H37Rv ----MIDRPLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICE-QIA
MUL_3392|M.ulcerans_Agy99 -MSRPLDGPLSGRVAFITGAARGQGRAHALRLARDGADVIAVDLCD-QIA
MAB_3081|M.abscessus_ATCC_1997 ------MGKLDGKVAFITGAARGQGRAHAIKLASEGASIIAVDICA-QIP
MMAR_4754|M.marinum_M -MTEQLGGRVAGKVAFITGAARGQGRSHAVRLAQEGADIIAVDVCKPIVE
TH_3237|M.thermoresistible__bu -----MAGRLEGKVAFITGAARGQGRAHAVRMAQEGADIIAVDICR-QID
MSMEG_5813|M.smegmatis_MC2_155 -----MSGRVAGKRVLVTGAARGMGRSHAVRLAEEGADLILLDICE-SIP
Mvan_3395|M.vanbaalenii_PYR-1 ----MSAGRVAGKRVLITGAARGMGRSHAVRLAGEGADLILVDICE-SLP
Mflv_1905|M.gilvum_PYR-GCK -----MSGRVEGKRVLITGAARGMGRSHAVRLAQEGADLILVDICE-SLP
MAV_3551|M.avium_104 MATDDATGRVAGKRVLITGAARGMGRSHAVRLAEQGADCILVDICC-TPT
MLBr_01740|M.leprae_Br4923 --MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRHC----
. *: ::**** * **: *:::* **. *
Mb2771|M.bovis_AF2122/97 SVPYPLSTADDLAATVELVEDAGGGIVAR-QGDVRDRASLSVALQAGLDE
Rv2750|M.tuberculosis_H37Rv SVPYPLSTADDLAATVELVEDAGGGIVAR-QGDVRDRASLSVALQAGLDE
MUL_3392|M.ulcerans_Agy99 SVPYPLGTAEELATTVKLVEDTGARIVAS-QADVRDREALAAALQAGIDE
MAB_3081|M.abscessus_ATCC_1997 SVEYPMATTEDLDLTVKEVDALGRAISAH-ETDVRDREALQAAFDAGVHD
MMAR_4754|M.marinum_M NTTIPASTPEDLAETADLVKGHNRRIFTA-EADVRDYDALKAAVDAGVDE
TH_3237|M.thermoresistible__bu SVQIPLASPEDLAETADLVKGLNRRIYTA-EVDVRDFDALKAAVDTGVEQ
MSMEG_5813|M.smegmatis_MC2_155 EIEYPLATEDELAETVRLVEEHGRHALAH-VVDVRDGEALSAAVTDAVAQ
Mvan_3395|M.vanbaalenii_PYR-1 EIEYPLASPDDLAETARLVEDIGRRAVTH-VVDVRDGAALATAVDDGVAR
Mflv_1905|M.gilvum_PYR-GCK EVEYPLAAVEELADTAELVAQHGRRAVTH-VVDVRDEQALSAAVDDGVAR
MAV_3551|M.avium_104 GLDYPLATEEDLNETVRLVEKHGRRAVPK-IVDVRDEAAMKAAVDAAVDE
MLBr_01740|M.leprae_Br4923 ---------DGLAQTVSEARALGAQVPEYRALDISDYDEVAAFGADIHAR
: * *. . . *: * : .
Mb2771|M.bovis_AF2122/97 FGRLDIVVANAGIAMMQ-----AGDDGWRDVIDVNLTGVFHTVQVAIPTL
Rv2750|M.tuberculosis_H37Rv FGRLDIVVANAGIAMMQ-----AGDDGWRDVIDVNLTGVFHTVQVAIPTL
MUL_3392|M.ulcerans_Agy99 LGQVDIVVANAGIAPMQ-----SGDDGWRDVIDVNLSGAYYTVEVAIPTM
MAB_3081|M.abscessus_ATCC_1997 LGPVDIVVANAGIATLTP---ECGDDGWRDVLDVNLTGVYHTIEVARPSM
MMAR_4754|M.marinum_M LGRLDIIVANAGIGNGGATLDKTSEHDWQEMIDVNLSGVWKSVKAAVPHL
TH_3237|M.thermoresistible__bu LGRLDIIVANAGIGNGGETLDKTSEGDWTDMIDVNLAGVWKTVKAGVPHI
MSMEG_5813|M.smegmatis_MC2_155 LGGLDASVANAGVLTAG-TWDTTTSEQWRTVVDVNLIGTWNTCAAALPHL
Mvan_3395|M.vanbaalenii_PYR-1 LGGLDAAVANAGVLTAG-TWDTTTAEQWRTVVDVNLIGTWNTCAAALPHL
Mflv_1905|M.gilvum_PYR-GCK LGGLDAAVANAGVLTGG-TWDTTTAQQWRTVIDVNLIGTWNTCRAAIPHL
MAV_3551|M.avium_104 LGGLDGAVANAGVLTVG-TWDTTTAEQWRLVLDVNLIGAWNTCAAALPHL
MLBr_01740|M.leprae_Br4923 HPSMDIVMNIAGVSVWG-TVDRLTHDQWSNVVAINLMGPIHIIETFVPAI
:* : **: * :: :** * . * :
Mb2771|M.bovis_AF2122/97 IEQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYANHLA
Rv2750|M.tuberculosis_H37Rv IEQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYANHLA
MUL_3392|M.ulcerans_Agy99 IEQGRGGSIVLISSAAGLVGISSADAGAIGYVASKHALVGLMRVYANLLA
MAB_3081|M.abscessus_ATCC_1997 VQRGAGGSIVLTSSVAGLTGMVMDNPGGLAYTAAKHGLVGLMRSYANMLA
MMAR_4754|M.marinum_M IAGGNGGSIVLTSSVGGMKAYPHCG----NYVAAKHGVVGLMRSFAVELG
TH_3237|M.thermoresistible__bu LAGGRGGSIILTSSVGGLKAYPHTG----HYVAAKHGVVGLMRTFAVELG
MSMEG_5813|M.smegmatis_MC2_155 VERGG--SVINISSAAGVKGSPLHTP----YTASKHGVVGLSRALANELA
Mvan_3395|M.vanbaalenii_PYR-1 VDRGG--SLVNISSAAGIKGTPLHTP----YTASKHGVVGMSKALANEVA
Mflv_1905|M.gilvum_PYR-GCK LERGG--SLINVSSAAGIKGSPLHTP----YTASKAGVVGMSKALANELA
MAV_3551|M.avium_104 IGGGGG-SLVNISSSAGIKGTPLHLP----YTASKHGIVGMTLALANELA
MLBr_01740|M.leprae_Br4923 LAAGRGGHLVNVSSAAGLVALPWHAA----YSASKYGLRGLSEVLRFDLA
: * :: ** .*: . * *:* .: *: :.
Mb2771|M.bovis_AF2122/97 PQNIRVNSVHPCGVDTPMINNEFFQQWLT--TADMDAP-----HNLGNAL
Rv2750|M.tuberculosis_H37Rv PQNIRVNSVHPCGVDTPMINNEFFQQWLT--TADMDAP-----HNLGNAL
MUL_3392|M.ulcerans_Agy99 PHSIRVNSLHPSGVDPPMINNEFIRHWLADLVAETGSG-----PGAGNAL
MAB_3081|M.abscessus_ATCC_1997 PHNIRVNTIHPSGVATPMLINPVTEKLFAD--GSPEGM-----SDTGNAL
MMAR_4754|M.marinum_M QHMIRVNSVHPTHVRTPMLHNEGTFKMFRPDLENPGPDDMAPICQLFHTL
TH_3237|M.thermoresistible__bu QHNIRVNSVHPTNVNTPLFMNEPTMKLFRPDLENPGPEDLKVVAQMMHTL
MSMEG_5813|M.smegmatis_MC2_155 AVGVRVNTVHPTGVETGMRP-DSLHTLLVEKRPDLAPI-------FANAM
Mvan_3395|M.vanbaalenii_PYR-1 AQSVRVNTVHPTGVETGMRP-ESLRDLIAQQRPDLGPL-------FLNAM
Mflv_1905|M.gilvum_PYR-GCK AMNIRVNTVHPTGVPTGMRP-ASLHQLLHDTRSDLSPI-------FQNAL
MAV_3551|M.avium_104 AQNIRVNTVHPTGVATGMAP-PGMHALIAEQRPDLVPI-------FLNAL
MLBr_01740|M.leprae_Br4923 RYRIGVSVVVPGAVRTPLVDTIEIAGVDRQDPRFSRWVDRFRGHAVSAER
: *. : * * . :
Mb2771|M.bovis_AF2122/97 PVELVQPTDIANAVAWLASEEARYVTGVT---------------------
Rv2750|M.tuberculosis_H37Rv PVELVQPTDIANAVAWLASEEARYVTGVT---------------------
MUL_3392|M.ulcerans_Agy99 PVQILQADDIAGALAWLVSDEARYITGVA---------------------
MAB_3081|M.abscessus_ATCC_1997 PVVLMEPEDMANTVAWLVSEDARYITGVA---------------------
MMAR_4754|M.marinum_M PIPWVEAEDISNAVLFLASDESRYITGVT---------------------
TH_3237|M.thermoresistible__bu PIGWVEPEDIANAVLFLASDEARYITGVT---------------------
MSMEG_5813|M.smegmatis_MC2_155 PIVMAEAIDISNAVLYLVSDESRYVTGLE---------------------
Mvan_3395|M.vanbaalenii_PYR-1 PIQMAEAVDVSNAVLFLVSDESRYVTGLE---------------------
Mflv_1905|M.gilvum_PYR-GCK PIVMAEAVDISNAVLFLISDEARHVTGLE---------------------
MAV_3551|M.avium_104 PAPLIEASDVSNAVLYLISDESRYVTGLE---------------------
MLBr_01740|M.leprae_Br4923 AAEKILAGVAKNRYLIYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVFTRA
. . . *.: * : *.
Mb2771|M.bovis_AF2122/97 -----------LPVDAGFVNKR----------
Rv2750|M.tuberculosis_H37Rv -----------LPVDAGFVNKR----------
MUL_3392|M.ulcerans_Agy99 -----------LPVDAGCVNKR----------
MAB_3081|M.abscessus_ATCC_1997 -----------LPVDAGFTNRR----------
MMAR_4754|M.marinum_M -----------LPVDAGGCLK-----------
TH_3237|M.thermoresistible__bu -----------LPVDGGSCLK-----------
MSMEG_5813|M.smegmatis_MC2_155 -----------FKVDAGVTLR-----------
Mvan_3395|M.vanbaalenii_PYR-1 -----------FKVDAGVTLR-----------
Mflv_1905|M.gilvum_PYR-GCK -----------FKVDAGVTIR-----------
MAV_3551|M.avium_104 -----------LKVDAGVTIR-----------
MLBr_01740|M.leprae_Br4923 LRPSPATMRCVEPIKVGVYSQPRSGEGGKSGN
:. * :