For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSAGRVAGKRVLITGAARGMGRSHAVRLAGEGADLILVDICESLPEIEYPLASPDDLAETARLVEDIGRR AVTHVVDVRDGAALATAVDDGVARLGGLDAAVANAGVLTAGTWDTTTAEQWRTVVDVNLIGTWNTCAAAL PHLVDRGGSLVNISSAAGIKGTPLHTPYTASKHGVVGMSKALANEVAAQSVRVNTVHPTGVETGMRPESL RDLIAQQRPDLGPLFLNAMPIQMAEAVDVSNAVLFLVSDESRYVTGLEFKVDAGVTLR
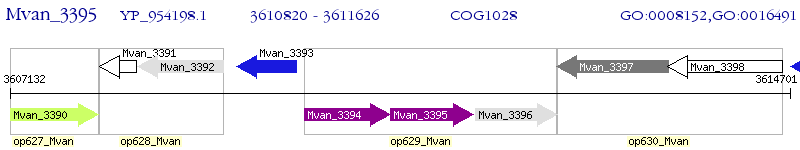
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3395 | - | - | 100% (268) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_5168 | - | 5e-64 | 50.00% (280) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_3394 | - | 9e-63 | 48.52% (270) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2771 | - | 5e-60 | 46.97% (264) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1905 | - | 1e-123 | 80.83% (266) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2750 | - | 5e-60 | 46.97% (264) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01740 | - | 7e-26 | 36.68% (199) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0073c | - | 3e-59 | 47.99% (273) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_4754 | - | 5e-63 | 49.82% (281) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_3551 | - | 1e-111 | 75.00% (268) | carveol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5813 | - | 1e-127 | 82.71% (266) | carveol dehydrogenase |
| M. thermoresistible (build 8) | TH_3757 | - | 4e-65 | 49.25% (268) | PUTATIVE carveol dehydrogenase |
| M. ulcerans Agy99 | MUL_3392 | fabG | 6e-58 | 44.85% (272) | 3-ketoacyl-(acyl-carrier-protein) reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2771|M.bovis_AF2122/97 ----MIDRPLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICEQIA-
Rv2750|M.tuberculosis_H37Rv ----MIDRPLEGKVAFITGAARGLGRAHAVRLAADGANIIAVDICEQIA-
MUL_3392|M.ulcerans_Agy99 -MSRPLDGPLSGRVAFITGAARGQGRAHALRLARDGADVIAVDLCDQIA-
TH_3757|M.thermoresistible__bu ------MGKLDGKVAFITGAARGQGRAHAVRLAEEGADVIAVDVCEQYA-
Mvan_3395|M.vanbaalenii_PYR-1 ----MSAGRVAGKRVLITGAARGMGRSHAVRLAGEGADLILVDICESLP-
MSMEG_5813|M.smegmatis_MC2_155 -----MSGRVAGKRVLVTGAARGMGRSHAVRLAEEGADLILLDICESIP-
Mflv_1905|M.gilvum_PYR-GCK -----MSGRVEGKRVLITGAARGMGRSHAVRLAQEGADLILVDICESLP-
MAV_3551|M.avium_104 MATDDATGRVAGKRVLITGAARGMGRSHAVRLAEQGADCILVDICCTPT-
MAB_0073c|M.abscessus_ATCC_199 --MTRLG----GRVAVVTGAGKGMGRTHCERLAEEGSDVIAID-------
MMAR_4754|M.marinum_M -MTEQLGGRVAGKVAFITGAARGQGRSHAVRLAQEGADIIAVDVCKPIVE
MLBr_01740|M.leprae_Br4923 --MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDR------
*: .:***. * **: . *** *:: *
Mb2771|M.bovis_AF2122/97 SVPYPLSTADDLAATVELVEDAGGGIVAR-QGDVRDRASLSVALQAGLDE
Rv2750|M.tuberculosis_H37Rv SVPYPLSTADDLAATVELVEDAGGGIVAR-QGDVRDRASLSVALQAGLDE
MUL_3392|M.ulcerans_Agy99 SVPYPLGTAEELATTVKLVEDTGARIVAS-QADVRDREALAAALQAGIDE
TH_3757|M.thermoresistible__bu TVVYQMSRPEDLAETVRQVETLGRRIVAR-EVDVCDLAGLQKAFDDGAAQ
Mvan_3395|M.vanbaalenii_PYR-1 EIEYPLASPDDLAETARLVEDIGRRAVTH-VVDVRDGAALATAVDDGVAR
MSMEG_5813|M.smegmatis_MC2_155 EIEYPLATEDELAETVRLVEEHGRHALAH-VVDVRDGEALSAAVTDAVAQ
Mflv_1905|M.gilvum_PYR-GCK EVEYPLAAVEELADTAELVAQHGRRAVTH-VVDVRDEQALSAAVDDGVAR
MAV_3551|M.avium_104 GLDYPLATEEDLNETVRLVEKHGRRAVPK-IVDVRDEAAMKAAVDAAVDE
MAB_0073c|M.abscessus_ATCC_199 ---LPGS--DDLAQTAAAVEEHGRRCVWA-FADVRDAAALTEAIDTGVRM
MMAR_4754|M.marinum_M NTTIPASTPEDLAETADLVKGHNRRIFTA-EADVRDYDALKAAVDAGVDE
MLBr_01740|M.leprae_Br4923 -------HCDGLAQTVSEARALGAQVPEYRALDISDYDEVAAFGADIHAR
: * *. . . *: * :
Mb2771|M.bovis_AF2122/97 FGRLDIVVANAGIAM-----MQAGDDGWRDVIDVNLTGVFHTVQVAIPTL
Rv2750|M.tuberculosis_H37Rv FGRLDIVVANAGIAM-----MQAGDDGWRDVIDVNLTGVFHTVQVAIPTL
MUL_3392|M.ulcerans_Agy99 LGQVDIVVANAGIAP-----MQSGDDGWRDVIDVNLSGAYYTVEVAIPTM
TH_3757|M.thermoresistible__bu LGGVDIVVANAGIGPG---GQTTDDEQWDDVVNVNLKGVWNTLRVAVPTM
Mvan_3395|M.vanbaalenii_PYR-1 LGGLDAAVANAGVLTA-GTWDTTTAEQWRTVVDVNLIGTWNTCAAALPHL
MSMEG_5813|M.smegmatis_MC2_155 LGGLDASVANAGVLTA-GTWDTTTSEQWRTVVDVNLIGTWNTCAAALPHL
Mflv_1905|M.gilvum_PYR-GCK LGGLDAAVANAGVLTG-GTWDTTTAQQWRTVIDVNLIGTWNTCRAAIPHL
MAV_3551|M.avium_104 LGGLDGAVANAGVLTV-GTWDTTTAEQWRLVLDVNLIGAWNTCAAALPHL
MAB_0073c|M.abscessus_ATCC_199 LGRLDIVVANAGVYDGLAPSCELTEDSWRRALDVNLTGAWHTVKAGAAHL
MMAR_4754|M.marinum_M LGRLDIIVANAGIGNGGATLDKTSEHDWQEMIDVNLSGVWKSVKAAVPHL
MLBr_01740|M.leprae_Br4923 HPSMDIVMNIAGVSVW-GTVDRLTHDQWSNVVAINLMGPIHIIETFVPAI
:* : **: . * : :** * . . :
Mb2771|M.bovis_AF2122/97 IEQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYANHLA
Rv2750|M.tuberculosis_H37Rv IEQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYANHLA
MUL_3392|M.ulcerans_Agy99 IEQGRGGSIVLISSAAGLVGISSADAGAIGYVASKHALVGLMRVYANLLA
TH_3757|M.thermoresistible__bu LTQDRGGSIILTSSTGGLIGTPVVSGGMIAYTAAKHGVIGIMRAYANYLA
Mvan_3395|M.vanbaalenii_PYR-1 VD-RGG-SLVNISSAAGIKGTPLHTP----YTASKHGVVGMSKALANEVA
MSMEG_5813|M.smegmatis_MC2_155 VE-RGG-SVINISSAAGVKGSPLHTP----YTASKHGVVGLSRALANELA
Mflv_1905|M.gilvum_PYR-GCK LE-RGG-SLINVSSAAGIKGSPLHTP----YTASKAGVVGMSKALANELA
MAV_3551|M.avium_104 IG-GGGGSLVNISSSAGIKGTPLHLP----YTASKHGIVGMTLALANELA
MAB_0073c|M.abscessus_ATCC_199 LD---GGSVIIISSTNGLKGTANTAH----YTASKHAVVGLARTAANELG
MMAR_4754|M.marinum_M IAGGNGGSIVLTSSVGGMKAYPHCGN----YVAAKHGVVGLMRSFAVELG
MLBr_01740|M.leprae_Br4923 LAAGRGGHLVNVSSAAGLVALPWHAA----YSASKYGLRGLSEVLRFDLA
: * :: ** *: . * *:* .: *: :.
Mb2771|M.bovis_AF2122/97 PQNIRVNSVHPCGVDTPMINNEFFQQWLT--TADMDAPH----------N
Rv2750|M.tuberculosis_H37Rv PQNIRVNSVHPCGVDTPMINNEFFQQWLT--TADMDAPH----------N
MUL_3392|M.ulcerans_Agy99 PHSIRVNSLHPSGVDPPMINNEFIRHWLADLVAETGSGP----------G
TH_3757|M.thermoresistible__bu PHHIRVNSVAPTTVATPMANNGDLSMLKK---YEPAIAR----------S
Mvan_3395|M.vanbaalenii_PYR-1 AQSVRVNTVHPTGVETGMRPESLRDLIAQ---QRPDLGP----------L
MSMEG_5813|M.smegmatis_MC2_155 AVGVRVNTVHPTGVETGMRPDSLHTLLVE---KRPDLAP----------I
Mflv_1905|M.gilvum_PYR-GCK AMNIRVNTVHPTGVPTGMRPASLHQLLHD---TRSDLSP----------I
MAV_3551|M.avium_104 AQNIRVNTVHPTGVATGMAPPGMHALIAE---QRPDLVP----------I
MAB_0073c|M.abscessus_ATCC_199 PQGIRVNTVHPGAVSTGMILN--PAVIAR---LRPDLEHPTVEDAAEALA
MMAR_4754|M.marinum_M QHMIRVNSVHPTHVRTPMLHN--EGTFKM---FRPDLENPGPDDMAPICQ
MLBr_01740|M.leprae_Br4923 RYRIGVSVVVPGAVRTPLVDTIEIAGVDR---QDPRFSRWVDRFRG--HA
: *. : * * . :
Mb2771|M.bovis_AF2122/97 LGNALPVELVQPTDIANAVAWLASEEARYVTGVT----------------
Rv2750|M.tuberculosis_H37Rv LGNALPVELVQPTDIANAVAWLASEEARYVTGVT----------------
MUL_3392|M.ulcerans_Agy99 AGNALPVQILQADDIAGALAWLVSDEARYITGVA----------------
TH_3757|M.thermoresistible__bu LENAIPVDYVEARDIANAVAWLASDDARYVTGTV----------------
Mvan_3395|M.vanbaalenii_PYR-1 FLNAMPIQMAEAVDVSNAVLFLVSDESRYVTGLE----------------
MSMEG_5813|M.smegmatis_MC2_155 FANAMPIVMAEAIDISNAVLYLVSDESRYVTGLE----------------
Mflv_1905|M.gilvum_PYR-GCK FQNALPIVMAEAVDISNAVLFLISDEARHVTGLE----------------
MAV_3551|M.avium_104 FLNALPAPLIEASDVSNAVLYLISDESRYVTGLE----------------
MAB_0073c|M.abscessus_ATCC_199 ARTLLPVPWVEAVDISNAVVFLASDEARYITGTQ----------------
MMAR_4754|M.marinum_M LFHTLPIPWVEAEDISNAVLFLASDESRYITGVT----------------
MLBr_01740|M.leprae_Br4923 VSAERAAEKILAGVAKNRYLIYTSHDIRVLYGFKRLAWWPYGVVMKQVNV
. . . *.: * : *
Mb2771|M.bovis_AF2122/97 ----------------LPVDAGFVNKR----------
Rv2750|M.tuberculosis_H37Rv ----------------LPVDAGFVNKR----------
MUL_3392|M.ulcerans_Agy99 ----------------LPVDAGCVNKR----------
TH_3757|M.thermoresistible__bu ----------------VPVDAGLLNKR----------
Mvan_3395|M.vanbaalenii_PYR-1 ----------------FKVDAGVTLR-----------
MSMEG_5813|M.smegmatis_MC2_155 ----------------FKVDAGVTLR-----------
Mflv_1905|M.gilvum_PYR-GCK ----------------FKVDAGVTIR-----------
MAV_3551|M.avium_104 ----------------LKVDAGVTIR-----------
MAB_0073c|M.abscessus_ATCC_199 ----------------LVVDAGLTQKA----------
MMAR_4754|M.marinum_M ----------------LPVDAGGCLK-----------
MLBr_01740|M.leprae_Br4923 VFTRALRPSPATMRCVEPIKVGVYSQPRSGEGGKSGN
:..* :