For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PPPLHSPAFYAGDPFPAYRELRATDPVTWNEEHGFWALLKYEDIRYVSTNPALFSSARGITVPDPAIENP VMEGSLIFTDPPRHRRLRKLINSGFTRRQVAILEPKLRAFARTALDAIDPAGGTEFAERIAAPLPTRMIA ELLGAPDEDWEQFRRWSDAAVGMDDPDVELDTFTAMGELYQYFEKLIALRRNGALRDSDDLLSVLVAAEV DGVRLSDQDLLQFCLLLLVAGNETTRNLIALGTLALIEHPQQFRLLREHPELIPTAVEEFLRYTSPVANM TRCATRDVEMRGRLIREGQYVTMLYGSANRDEDIFGPTSEQLDVTRNPNPHLAFGCGEHSCLGAQLARLE ARVLFEELLARFEHIEPAGEVLRMQATMVPGVRAMPVRLRAATDPRRAKARAG*
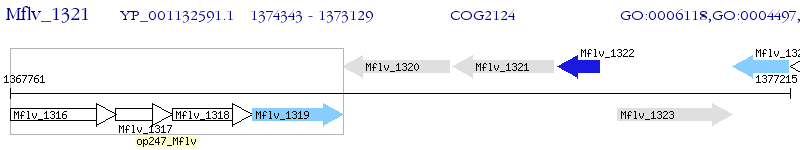
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1321 | - | - | 100% (404) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_0425 | - | 1e-77 | 43.77% (393) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_3307 | - | 1e-70 | 40.85% (399) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2289 | cyp124 | 4e-67 | 40.28% (360) | cytochrome P450 124 CYP124 |
| M. tuberculosis H37Rv | Rv2266 | cyp124 | 4e-67 | 40.28% (360) | cytochrome P450 124 CYP124 |
| M. leprae Br4923 | MLBr_02088 | - | 1e-50 | 33.25% (391) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_1406c | - | 3e-60 | 37.67% (369) | cytochrome P450 |
| M. marinum M | MMAR_5003 | cyp142A3 | 7e-65 | 39.37% (381) | cytochrome P450 142A3 Cyp142A3 |
| M. avium 104 | MAV_1339 | - | 1e-158 | 70.18% (389) | cytochrome P450 superfamily protein |
| M. smegmatis MC2 155 | MSMEG_3551 | - | 6e-72 | 40.92% (391) | linalool 8-monooxygenase |
| M. thermoresistible (build 8) | TH_2649 | - | 1e-151 | 68.38% (389) | PUTATIVE cytochrome P450 superfamily protein |
| M. ulcerans Agy99 | MUL_1282 | cyp124A1 | 1e-64 | 39.00% (359) | cytochrome P450 124A1, Cyp124A1 |
| M. vanbaalenii PYR-1 | Mvan_5481 | - | 0.0 | 85.89% (404) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1321|M.gilvum_PYR-GCK -------------------------------------MPPPLHSP---AF
Mvan_5481|M.vanbaalenii_PYR-1 ----------------------------------MSRLSNPLHSP---AF
MAV_1339|M.avium_104 ----------------------------MQTHPPLRSPSFPLHSP---DF
TH_2649|M.thermoresistible__bu -----------------------------------VLTTVPLHSP---DF
Mb2289|M.bovis_AF2122/97 ----MGLNTAIATRVNGTPPPEVPIADIELGSLDFWALDDDVRDG---AF
Rv2266|M.tuberculosis_H37Rv ----MGLNTAIATRVNGTPPPEVPIADIELGSLDFWALDDDVRDG---AF
MUL_1282|M.ulcerans_Agy99 MDLSTNLNTGLLPRVNGTPPPEVPLADIELGSLEFWGRDDDFRDG---AF
MSMEG_3551|M.smegmatis_MC2_155 --------MGIAPRVNGAAPPDVPLSEINLGSWDFWALDDDIRDG---AF
MMAR_5003|M.marinum_M ------------------------------MTKPLIKPDVDLTDG---NF
MAB_1406c|M.abscessus_ATCC_199 -----------------------------MSVDVSHTVAPRYVPG---TA
MLBr_02088|M.leprae_Br4923 ------MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDP
Mflv_1321|M.gilvum_PYR-GCK YAG-DPFPAYRELRATDPVT-----WNEEHGFWALLKYEDIRYVSTNPAL
Mvan_5481|M.vanbaalenii_PYR-1 YAG-DPFPVYRELRATDPVC-----WNDEHEFWALLKYEDIRYVSTNPAL
MAV_1339|M.avium_104 YAG-NPYPAYRELRATAPVC-----WNDVTNFWALLKYEDIRFVSSNPAL
TH_2649|M.thermoresistible__bu YAG-NPYPAYRELRATTPVV-----YNDVTNFWALLKYEDIRYVSSHPQL
Mb2289|M.bovis_AF2122/97 ATLRREAPISFWPTIELPG------FVAGNGHWALTKYDDVFYASRHPDI
Rv2266|M.tuberculosis_H37Rv ATLRREAPISFWPTIELPG------FVAGNGHWALTKYDDVFYASRHPDI
MUL_1282|M.ulcerans_Agy99 ATLRREAPISFWPPIELAG------LTAGKGHWALTKHDDIHFASRHPEI
MSMEG_3551|M.smegmatis_MC2_155 ATLRREAPISFHPAFDTEEG-----FPQGAGHWALTRHDDVFYASRHPEL
MMAR_5003|M.marinum_M YASRQAREAYRWMRANQPV------FRDRNGLAAASTYQAVIDAERQPEL
MAB_1406c|M.abscessus_ATCC_199 DNWTNPWPMYAALRDHDPVHHVVPEYTPDQDYYVLTRHADVYEAARDWET
MLBr_02088|M.leprae_Br4923 GTRADPFPVYRALIDYGPMQ------LPGMPLTVFSSFSDCDEALRHPLS
. . . .
Mflv_1321|M.gilvum_PYR-GCK FSSARGITVPDPAIENPVMEGS-----LIFTDPPRHRRLRKLINSGFTRR
Mvan_5481|M.vanbaalenii_PYR-1 FSSARGVTVPDPALENPVMEGS-----LIFTDPPRHRQLRKLINSGFTRR
MAV_1339|M.avium_104 FTSTRGITIPDPQLPNPVQQGS-----LIFTDPPRHRQLRKLINSGFTRR
TH_2649|M.thermoresistible__bu FSSTKGITIPDPDQPEPVQEGN-----LIFTDPPRHRQLRKLINTAFTRR
Mb2289|M.bovis_AF2122/97 FSSYPNITINDQTPELAEYFGS-----MIVLDDPRHQRLRSIVSRAFTPK
Rv2266|M.tuberculosis_H37Rv FSSYPNITINDQTPELAEYFGS-----MIVLDDPRHQRLRSIVSRAFTPK
MUL_1282|M.ulcerans_Agy99 FHSSPNIVIHDQTPELAEYFGS-----MIVLDDPRHQRLRSIVSRAFTPK
MSMEG_3551|M.smegmatis_MC2_155 FSSASGIVIGDQTPELAEYFGS-----MIAMDDPRHTRLRNIVRSAFTPR
MMAR_5003|M.marinum_M FSNAGGIRPD------QDALPM-----MIDMDDPAHLLRRKLVNAGFTRK
MAB_1406c|M.abscessus_ATCC_199 FSSARGLTVTYGDLEKTGMGDNP---PMVMQDPPTHTEFRKLVSRGFTPR
MLBr_02088|M.leprae_Br4923 ASDRLKATLAQQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAPK
. :: * * * *.:: .*: :
Mflv_1321|M.gilvum_PYR-GCK QVAILEPKLRAFARTALDAIDPAG---GTEFAERIAAPLPTRMIAELLGA
Mvan_5481|M.vanbaalenii_PYR-1 QVAILEPKLRAFARAALDAIDPS----ATEFAEQVAAPLPTRMIAELLGA
MAV_1339|M.avium_104 RVSVLEPKIREIVRGILDGIERGA---VHEFAEQIAAPLPTRMIAELIGA
TH_2649|M.thermoresistible__bu QVALLEPKVREIVYGIFEGVAPGS---EQEFAESFAAPLPTRMIAELLGA
Mb2289|M.bovis_AF2122/97 VVARIEAAVRDRAHRLVSSMIANNPDRQADLVSELAGPLPLQIICDMMGI
Rv2266|M.tuberculosis_H37Rv VVARIEAAVRDRAHRLVSSMIANNPDRQADLVSELAGPLPLQIICDMMGI
MUL_1282|M.ulcerans_Agy99 VVARIEASVRERAHRLVAAMIENHPDGQADLVSELAGPLPLQIICDMMGI
MSMEG_3551|M.smegmatis_MC2_155 VLAVIEDSVRDRARRLVAGMVDRNPDGHAELVTELAGPLPLQIICDMMGI
MMAR_5003|M.marinum_M RVKDKERSIAQLCDTLIDAVCER---GECDFVRDLAAPLPMAVIGDMLGV
MAB_1406c|M.abscessus_ATCC_199 QVTAVEPKVREFVVQRLERLKERG---EGDIVVELFKPLPSMVVAHYLGV
MLBr_02088|M.leprae_Br4923 VVQALEGDIAALVDSLLDKGAAAG---QFDVIADLAFPLAVAVICRLLGV
: * : . :. . **. :: :*
Mflv_1321|M.gilvum_PYR-GCK PDEDWEQFRRWSDAAVGMDDP-----DVEL----DTFTAMGELYQYFEKL
Mvan_5481|M.vanbaalenii_PYR-1 PDEDWEQFRTWSDATVGMDDP-----DVEL----DTFAAMGELYQYFEAQ
MAV_1339|M.avium_104 PPDDWEQFRAWSDAATGTADP-----EIEL----DPAVAAGQLYEYFQRL
TH_2649|M.thermoresistible__bu SPEDWERFRRWSDACTGTADP-----EIEL----DAMEAIGELYEYFQQL
Mb2289|M.bovis_AF2122/97 PKADHQRIFHWTNVILGFGDP-----DLAT-DFDEFMQVSADIGAYATAL
Rv2266|M.tuberculosis_H37Rv PKADHQRIFHWTNVILGFGDP-----DLAT-DFDEFMQVSADIGAYATAL
MUL_1282|M.ulcerans_Agy99 PEEDHEQIFHWTNVILGFGDP-----DLTT-DFDDFLQVSMAIGGYATAL
MSMEG_3551|M.smegmatis_MC2_155 PESDHQQIFHWTNVILGFGDP-----DLTT-DFDEFVTVAMDIGAYATAL
MMAR_5003|M.marinum_M LPEQREMFLRWSDDLVTFLSS-----HVSQEDFQVTIDAFAAYNDFTRAT
MAB_1406c|M.abscessus_ATCC_199 PEEDRTQFDAWTDGIVAAASGGA--IDIEAMQG-EVAQTIGELMMYFTGL
MLBr_02088|M.leprae_Br4923 PYEDAPEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQL
: : : . . :
Mflv_1321|M.gilvum_PYR-GCK IALRRNGALRDSDDLLSVLVAAEV--DGVRLSDQDLLQFCLLLLVAGNET
Mvan_5481|M.vanbaalenii_PYR-1 IARRRDGTLRDSDDLLSVLVAAEV--DGVRLTDQDLLQFCLLLLVAGNET
MAV_1339|M.avium_104 IAARR---ARPRADLLSVLAEAEI--DEHRLTDEDLLNFAFLLLVAGNET
TH_2649|M.thermoresistible__bu IAARR---TEPRDDMMSVLAQAEV--DGARLSDEDLLNFAFLLLVAGNET
Mb2289|M.bovis_AF2122/97 AEDRR---VNHHDDLTSSLVEAEV--DGERLSSREIASFFILLVVAGNET
Rv2266|M.tuberculosis_H37Rv AEDRR---VNHHDDLTSSLVEAEV--DGERLSSREIASFFILLVVAGNET
MUL_1282|M.ulcerans_Agy99 ADDRR---VNHHGDLTTSLVEAEV--DGERLSSSEIAMFFILLVVAGNET
MSMEG_3551|M.smegmatis_MC2_155 ADQRR---SAPADDLTTSLVQAEV--DGERLTSAEVASFFILLVVAGNET
MMAR_5003|M.marinum_M IAARR---AEPTDDLVSVLVSSEV--DGERLSDDELVMETLLILIGGDET
MAB_1406c|M.abscessus_ATCC_199 IERRR---AEPEDDTVSHLVAAGVGADGDISGTLQILGFAFTMVTGGNDT
MLBr_02088|M.leprae_Br4923 VKCRR---GTPGEDLISRLIELDE--SGDQLTEEEIIATCGLLLVAGHET
** * : * . :: :: .*.:*
Mflv_1321|M.gilvum_PYR-GCK TRNLIALGTLALIEHPQQFRLLREHPE-LIPTAVEEFLRYTSPVANMTRC
Mvan_5481|M.vanbaalenii_PYR-1 TRNLVALGTLALIENPDQFRLLRQRRD-LVPTAVEEMLRYASPVANMTRC
MAV_1339|M.avium_104 TRNLIALGTLALIAHPDQYRLLVEEPA-RIPLAVEEMLRWNSPVVHMART
TH_2649|M.thermoresistible__bu TRNLIALGTLALIEHPDQCRKLIENPT-LIPGAVEEMLRYTSPVTHMARQ
Mb2289|M.bovis_AF2122/97 TRNAITHGVLALSRYPEQRDRWWSDFDGLAPTAVEEIVRWASPVVYMRRT
Rv2266|M.tuberculosis_H37Rv TRNAITHGVLALSRYPEQRDRWWSDFDGLAPTAVEEIVRWASPVVYMRRT
MUL_1282|M.ulcerans_Agy99 TRNTISHGMLALSRYPDERAKWWSDFDGLAATAVEEIVRWASPVVYMRRT
MSMEG_3551|M.smegmatis_MC2_155 TRNAISHGVLALTRYPEQRRLWWSRFDELAPTAVEEIVRWASPVSYMRRT
MMAR_5003|M.marinum_M TRHTLSGGSEQLLRNRDQWDLLQSDRE-LLPGAIEEMLRWTAPVKNMCRM
MAB_1406c|M.abscessus_ATCC_199 TTGMLGGAIQLLHQNPAQRQKLIDDPS-LIPGAVEEFLRLTSPVQGLART
MLBr_02088|M.leprae_Br4923 TVNLIANAVLAMLRNPSQWKALSSNPQ-RAPLVVEETLRYDPAIHLIGRV
* : . : : . . .:** :* ..: : *
Mflv_1321|M.gilvum_PYR-GCK ATRDVEMRGRLIREGQYVTMLYGSANRDEDIFGPTSEQLDVTRNPNPHLA
Mvan_5481|M.vanbaalenii_PYR-1 ATRDVDIRGRSIRAGQYVTMLYGSANRDKDVFGPTSEQFDITRNPNPHLA
MAV_1339|M.avium_104 ATADVEIRGQRIRAGEVVVMLYGSANRDEDVFGPDSEEFDVTRHPNPHIA
TH_2649|M.thermoresistible__bu ATEDVEIRGRHIRKGDTVVMLYGSANRDEEIFGDDAEDFRIDRQPNPHIA
Mb2289|M.bovis_AF2122/97 LTQDIELRGTKMAAGDKVSLWYCSANRDESKFAD-PWTFDLARNPNPHLG
Rv2266|M.tuberculosis_H37Rv LTQDIELRGTKMAAGDKVSLWYCSANRDESKFAD-PWTFDLARNPNPHLG
MUL_1282|M.ulcerans_Agy99 LNQDVDLRGTKMAAGDKVTLWYCSANRDEEKFAD-PWTFDVTRNPNPQVG
MSMEG_3551|M.smegmatis_MC2_155 VTRDTELAGTALPAGSKVTLWYGSANRDETKFDN-PWMFDVQRHPNPHVG
MMAR_5003|M.marinum_M LTADTEFHGTALSEGEKIMLLFESANFDEAVFTD-PEKFDIQRNPNSHLA
MAB_1406c|M.abscessus_ATCC_199 ATRDVIIGDTTIPAGRRALLLYGSANRDEREYGNDAAELDVLRKPRNILT
MLBr_02088|M.leprae_Br4923 AAKDMTIGQTTLTEGDTMVLLLAAANRDPAVYSR-PDEFDPDRPSSRHLA
* : : * : :** * : . : * . :
Mflv_1321|M.gilvum_PYR-GCK FGCG-EHSCLGAQLARLEARVLFEELLARFEHIEPAGEVLRMQA---TMV
Mvan_5481|M.vanbaalenii_PYR-1 FGCG-EHSCLGAQLARLEARVLFEELLNRFDHIELDGDVLRMQA---TMV
MAV_1339|M.avium_104 FGCG-EHSCVGAQLARLEATVFFEELLRRYPRIELVGEVDRMRA---TMV
TH_2649|M.thermoresistible__bu FGCG-EHSCIGAQLARLEACVFFEVLLGRFPDLELVGTVDRMRA---TMV
Mb2289|M.bovis_AF2122/97 FGGGGAHFCLGANLARREIRVAFDELRRQMPDVVATEEPARLLS---QFI
Rv2266|M.tuberculosis_H37Rv FGGGGAHFCLGANLARREIRVAFDELRRQMPDVVATEEPARLLS---QFI
MUL_1282|M.ulcerans_Agy99 FGGGGAHFCLGANLARREIRVVFDELRRQMPDVVATEEPARLLS---QFI
MSMEG_3551|M.smegmatis_MC2_155 FGGGGAHFCLGANLARREITVLFSELHRHLPDIVATEEPDRLQS---AFI
MMAR_5003|M.marinum_M FGFG-THFCMGNQLARLELSLMTARVVQRLPDLRLADQDSRLPLRPANFV
MAB_1406c|M.abscessus_ATCC_199 FSHG-NHHCLGAAAARMQSRIALEELLTRIPDFEVDLDGVTWADG--SYV
MLBr_02088|M.leprae_Br4923 FAVG-SHFCLGAALARLEATVTLSAISARFPQVQLAGELVYKPN---VAM
*. * * *:* ** : : : : . :
Mflv_1321|M.gilvum_PYR-GCK PGVRAMPVRLRAATDPRRAKARAG---
Mvan_5481|M.vanbaalenii_PYR-1 PGVRQMPVRLSATAAEPRETGTAAVSV
MAV_1339|M.avium_104 PGVKRMPVRMGA---------------
TH_2649|M.thermoresistible__bu PGVKRMPVRLGAGR-------------
Mb2289|M.bovis_AF2122/97 HGIKTLPVTWS----------------
Rv2266|M.tuberculosis_H37Rv HGIKTLPVTWS----------------
MUL_1282|M.ulcerans_Agy99 HGIKRLPVAWSR---------------
MSMEG_3551|M.smegmatis_MC2_155 HGIKRLPVAWR----------------
MMAR_5003|M.marinum_M SGLESMPVVFTPSRPLS----------
MAB_1406c|M.abscessus_ATCC_199 RRPLTVPIRVR----------------
MLBr_02088|M.leprae_Br4923 RGMSALPVQV-----------------
:*: