For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GEITPLDVVSGIDSVWDSVPHPVLVVDRGGMVRTLSASARSVLPDIGPGSAVEDFVPSWLSGDDDREART TELPDGEMAWWLLEDSARSTRLALTLERERSEFLDEASAVLMASLNIDRCMEATVQMAARHLADAAVLVA PASGGVVPLVFGLPDGTTEHRRIDADPAEVAGLSEALRGFPPVPSRWIDPGSLPEWLVPPGFSDGTGGVV ITPLPGHGVPAGALVLLRRSSESTFSDGEEVFARLFAARAGAALSAARLYAEQSTITRTLMRELLPPQLQ RLDGIELAGGYRASADHETIGGDFYDVHPAAGPDDETLVILGDVCGKGLDAAVLTGKIRNTLQALAPLAA DHTGVLKLLNSALLSIDDTRFATLVLASVVRRDGFVRLRLTSAGHLAPLIVRNDGSVEEADTGGTLVGVM PTIRARTVDAALAPGETCLLFTDGVTEARGGPLGADMFGEARLSAALSQCAGLPAEAVVERIMMLTSEWV HNRAHDDIAVVAITAPRRTHLSAVDGHTAGRFTA*
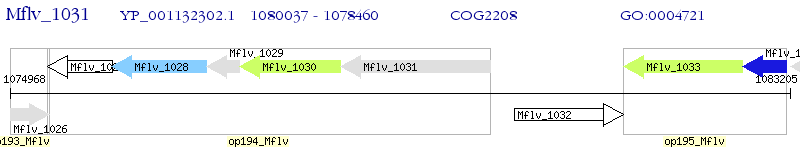
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1031 | - | - | 100% (525) | putative GAF sensor protein |
| M. gilvum PYR-GCK | Mflv_3857 | - | 1e-26 | 32.30% (291) | putative PAS/PAC sensor protein |
| M. gilvum PYR-GCK | Mflv_2742 | - | 2e-26 | 36.13% (238) | protein phosphatase 2C domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1399c | - | 2e-13 | 24.64% (276) | hypothetical protein Mb1399c |
| M. tuberculosis H37Rv | Rv1364c | - | 2e-13 | 24.64% (276) | hypothetical protein Rv1364c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_5186 | - | 1e-16 | 26.21% (309) | sensor-component of a two-component regulator |
| M. avium 104 | MAV_2108 | - | 3e-18 | 25.06% (423) | stage II sporulation protein E (SpoIIE) |
| M. smegmatis MC2 155 | MSMEG_6131 | - | 3e-14 | 30.34% (267) | magnesium or manganese-dependent protein phosphatase |
| M. thermoresistible (build 8) | TH_3243 | - | 5e-54 | 36.70% (406) | PUTATIVE putative putative GAF sensor protein |
| M. ulcerans Agy99 | MUL_3853 | rsbU | 3e-16 | 27.51% (269) | regulator of sigma subunit, anti-anti-sigma factor RsbU |
| M. vanbaalenii PYR-1 | Mvan_5805 | - | 0.0 | 76.89% (541) | putative GAF sensor protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1399c|M.bovis_AF2122/97 ------------------------MAAEMDWDKTVGAAEDVRRIFEHIPA
Rv1364c|M.tuberculosis_H37Rv ------------------------MAAEMDWDKTVGAAEDVRRIFEHIPA
MUL_3853|M.ulcerans_Agy99 MLLLDLLRQRTYSFVRSVHRRDGYMAAEMDWDKNVGTANTVRRIFENVPT
Mflv_1031|M.gilvum_PYR-GCK ----------------------------MGEITPLDVVSGIDSVWDSVPH
Mvan_5805|M.vanbaalenii_PYR-1 -------------------MWRGCEEATMGAGPPLDVDSGNDAAWAAVPH
TH_3243|M.thermoresistible__bu --------------------------------------------------
MAV_2108|M.avium_104 ----------MHTRLDELMAARDQLEQLVRVIAEIGSDLDLDVTLRRVLK
MMAR_5186|M.marinum_M ----------------------------MTETGDFGAVDAFQTQYADALR
MSMEG_6131|M.smegmatis_MC2_155 --------------------------------------------------
Mb1399c|M.bovis_AF2122/97 ILVGLEGPDHRFVAVNAAYRGFSPLLDT----------------------
Rv1364c|M.tuberculosis_H37Rv ILVGLEGPDHRFVAVNAAYRGFSPLLDT----------------------
MUL_3853|M.ulcerans_Agy99 MLVGLEGPEHRFVAANAAYRRFSPTFTA----------------------
Mflv_1031|M.gilvum_PYR-GCK PVLVVDR-GGMVRTLSASARSVLPDIGP----------------------
Mvan_5805|M.vanbaalenii_PYR-1 PVLVVDR-DGIVRTSSGSARSLLPGLSP----------------------
TH_3243|M.thermoresistible__bu -------------------------VSA----------------------
MAV_2108|M.avium_104 AAMELTGARYAVLSSRASDGGLLSFVHAGLDADTARQLGELAVGDGLRID
MMAR_5186|M.marinum_M AYLDTRDENSLAVGHEFGRRALQEQISILDIIEN----------------
MSMEG_6131|M.smegmatis_MC2_155 --------------------------------------------------
Mb1399c|M.bovis_AF2122/97 VGQPAREVYPELEGQQIYEMLDRVYQTGEPQSGSEWRLQTDYDGSGVEER
Rv1364c|M.tuberculosis_H37Rv VGQPAREVYPELEGQQIYEMLDRVYQTGEPQSGSEWRLQTDYDGSGVEER
MUL_3853|M.ulcerans_Agy99 VGQPAREVYPELESQQIYHMLDRVYETGEPQSGAEWRLQADFDGSGIDER
Mflv_1031|M.gilvum_PYR-GCK GSAVEDFVPSWLSGDD-------------DREARTTELPDGEMAWWLLED
Mvan_5805|M.vanbaalenii_PYR-1 GVALKDLAPSWLSGAHQHLTAASGSVHGRDFDAHPTPLPGGEVAWWLVED
TH_3243|M.thermoresistible__bu HTDIGDTAP-----------------------------------------
MAV_2108|M.avium_104 DVSVDPRSAHLSAHDPPLRALLGIPITVRAANFGNLYLADDRQGRVFTDS
MMAR_5186|M.marinum_M HFRLLDEISPNPGSDRSVALEFLLQTLAALDVATRGFLDGTKRYEEQRAR
MSMEG_6131|M.smegmatis_MC2_155 --------------------------------------------------
Mb1399c|M.bovis_AF2122/97 YFDFVVTPRRRADGSIEGVQ---LIVDDVTSRVRARQAAEARVEELSERY
Rv1364c|M.tuberculosis_H37Rv YFDFVVTPRRRADGSIEGVQ---LIVDDVTSRVRARQAAEARVEELSERY
MUL_3853|M.ulcerans_Agy99 YFDFLVTPRRREDGSIEGVQ---ILFDDVTTRVRARLAAEARVEELSERY
Mflv_1031|M.gilvum_PYR-GCK S---ARSTRLALTLERERSE---FLDEASAVLMASLNIDRCMEATVQMAA
Mvan_5805|M.vanbaalenii_PYR-1 SDRSARDARRALTVERERAA---FLDEASAVLMASLNIDRCMEATVQMAA
TH_3243|M.thermoresistible__bu --------------ASMGES---VIADVAETLGGSLNLRRTALRLISLIQ
MAV_2108|M.avium_104 QEGAVRAMATAAAAAIDNAR---LFERERESAKWTKASREITTALLSGDP
MMAR_5186|M.marinum_M AEDLAHRDKFRSALVNSLQEGFFVADDEGAVIDMNSAFAEILGYPPEGLP
MSMEG_6131|M.smegmatis_MC2_155 ----------------MRAP----FDRVIASPVDLTGPGQRRLSLLLVED
.
Mb1399c|M.bovis_AF2122/97 RN------VRDSATVMQQALLAASVPVVPGADIAAEYLVAAEDTAAGGDW
Rv1364c|M.tuberculosis_H37Rv RN------VRDSATVMQQALLAASVPVVPGADIAAEYLVAAEDTAAGGDW
MUL_3853|M.ulcerans_Agy99 RH------LRDSAIVMQQALLAPSVPVVSGPDIAAQYLVAAQDTAAGGDW
Mflv_1031|M.gilvum_PYR-GCK RH------LADAAVLVAP-ASGGVVPLVFGLPDGTTEHRRIDADPA----
Mvan_5805|M.vanbaalenii_PYR-1 RH------LADAAVLVTP-VAGGRIPVVCSGTDGTAEHRRVDADPS----
TH_3243|M.thermoresistible__bu PR------LADWAMVVLPDPRAGVLVAVGDAAPAGVPLRRADLDDL----
MAV_2108|M.avium_104 QTGPLQLIVNRALELAGAEQAILLVPREPESPGREADTLVVSATAGRYAS
MMAR_5186|M.marinum_M YRWPHPWLVDRKSAAQQQERVRHEGSADYETPVRHQDGHLSWVAVSINAV
MSMEG_6131|M.smegmatis_MC2_155 DR-------ADAILVEELIADAPDIDFVWAKSMSDAERTLADHRPD----
Mb1399c|M.bovis_AF2122/97 FDALALGDRLVLVVGDVVGHGVEAAAVMSQLRTALRMQISTGYTVVEALE
Rv1364c|M.tuberculosis_H37Rv FDALALGDRLVLVVGDVVGHGVEAAAVMSQLRTALRMQISAGYTVVEALE
MUL_3853|M.ulcerans_Agy99 FDAVALGDRLVLIVGDVVGHGVEAAAVMSQLRTALRLQIAAGRSIGEALE
Mflv_1031|M.gilvum_PYR-GCK --EVAGLSEALRGFPPVPSRWIDPGSLPEWLVPPG-FSDGTGGVVITPLP
Mvan_5805|M.vanbaalenii_PYR-1 --DVAGLSEALRGFPPVPSRWIDPASVPGWLVPES-FPGPVGSVVITPLP
TH_3243|M.thermoresistible__bu --PLGRVLRSGS-VEQIHDTDLAGLIPVEPFQQSA-VALAPAEVVAVGLT
MAV_2108|M.avium_104 QVIGRQVPMDGSTTGGVARRGLPLITDSFQYPIEGFTDVGERSAIVIPLI
MMAR_5186|M.marinum_M RGSGSDRDVYVGTVRDITAERAFAARESAVLRLATSVSVAKSVSEVLSIT
MSMEG_6131|M.smegmatis_MC2_155 -----CVLLDLNLPDAAGIDALHHLGKLDARIPIIVLTGLNDEHFGVSAV
Mb1399c|M.bovis_AF2122/97 AVDRFHKQVPGSKSATMCVGSLDFTSGEFQYCTAGHPPPLLVTADAS---
Rv1364c|M.tuberculosis_H37Rv AVDRFHKQVPGSKSATMCVGSLDFTSGEFQYCTAGHPPPLLVTADAS---
MUL_3853|M.ulcerans_Agy99 ALDAFHEHVPGSNSATLCVGSLDFTTGEFQYCTAGHPPPLVVTAEAK---
Mflv_1031|M.gilvum_PYR-GCK GHG----VPAGALVLLRRSSESTFSDGEEVFARLFAARAGAALSAAR---
Mvan_5805|M.vanbaalenii_PYR-1 GHG----VAAGALVLLRRTSQTAFSEGEEVFARLFAARAGAALSAAR---
TH_3243|M.thermoresistible__bu ARG----STFGALVVLRR-GDTGFRADELDVVARVADRAAVAFDSAR---
MAV_2108|M.avium_104 ADG----AVLGVIAVARDPQQPPFGNDYLDLVSDFARHAAIALALAAGRQ
MMAR_5186|M.marinum_M LEECR--TAVDVQRVVAVMWPPSDGEPTVQVAGEPAESSWRALDPQLR--
MSMEG_6131|M.smegmatis_MC2_155 ASG-------AQDYLVKGRVEPEMLRRAVLYAIERKRAELTSVDLHAS--
Mb1399c|M.bovis_AF2122/97 ---------------------------------ARYVEPTGAGPLGSGTG
Rv1364c|M.tuberculosis_H37Rv ---------------------------------ARYVEPTGAGPLGSGTG
MUL_3853|M.ulcerans_Agy99 ---------------------------------ARYLEPSGAGPLGSDTG
Mflv_1031|M.gilvum_PYR-GCK ---------------------------------LYAEQSTITRTLMRELL
Mvan_5805|M.vanbaalenii_PYR-1 ---------------------------------LYAEQSAITRTLLRDLL
TH_3243|M.thermoresistible__bu ---------------------------------LYEEQARVASVLQQSLH
MAV_2108|M.avium_104 HALNQELAQADTVDEAVRAAAEELRRLWRARRVLAVTFPSHTSATETAFG
MMAR_5186|M.marinum_M --------------------------------ETFLGARAQLPLTARPIE
MSMEG_6131|M.smegmatis_MC2_155 ----------------------------------QLRAQENARLERGLLP
Mb1399c|M.bovis_AF2122/97 FPVRSEVLN----------IGDAILFYTDGLIERPGRPLEAST-------
Rv1364c|M.tuberculosis_H37Rv FPVRSEVLN----------IGDAILFYTDGLIERPGRPLEAST-------
MUL_3853|M.ulcerans_Agy99 FPVRTELIG----------VGDVVLLYTDGLIERPGRPLGAST-------
Mflv_1031|M.gilvum_PYR-GCK PPQLQRLDG----------IELAGGYRASADHETIG--------------
Mvan_5805|M.vanbaalenii_PYR-1 PPQLHRLDG----------IELAGGYRASEDHEAVG--------------
TH_3243|M.thermoresistible__bu PPSLPDIPG----------VRLAACYRPAAEHLDIG--------------
MAV_2108|M.avium_104 PPEVVSVGEPTQWADLPSHMHQALGSLRDGDLLTPNTTQPGTAGIALQHP
MMAR_5186|M.marinum_M WPDSPGRAQG--------LVAVLSGTADVALWLELGAPRWVTAEDRLLVT
MSMEG_6131|M.smegmatis_MC2_155 SPLLMEGSG----------VDIVVEARPSRQHALIG--------------
* : .
Mb1399c|M.bovis_AF2122/97 -------AEFADLAASIASGSGGFVLDAPAR-------------------
Rv1364c|M.tuberculosis_H37Rv -------AEFADLAASIASGSGGFVLDAPAR-------------------
MUL_3853|M.ulcerans_Agy99 -------AEFADLAANITSG-GGFVIDAQSR-------------------
Mflv_1031|M.gilvum_PYR-GCK -------GDFYDVHPAAGPDDETLVILG----------------------
Mvan_5805|M.vanbaalenii_PYR-1 -------GDFYDVHPAATPDDETLVVLG----------------------
TH_3243|M.thermoresistible__bu -------GDFYDVT---GSDHDWLIAIG----------------------
MAV_2108|M.avium_104 DGVLVVWIDLTDERSFTLEDQTLLTVLAGRLGQGLQRVHQVDQQRETALA
MMAR_5186|M.marinum_M VLVGHLSLAIGHVRQFESARETSLTLQRAMLP------------------
MSMEG_6131|M.smegmatis_MC2_155 -------GDFYDVVQTPDRTVHVMIG------------------------
: . :
Mb1399c|M.bovis_AF2122/97 ---------PIDRLCSDTLELLLRSTGYNDDVTLLAMQRRAPTPPLHITL
Rv1364c|M.tuberculosis_H37Rv ---------PIDRLCSDTLELLLRSTGYNDDVTLLAMQRRAPTPPLHITL
MUL_3853|M.ulcerans_Agy99 ---------PIDRLCSETLELLLRSTGYNDDITLLAVQRRAPTPPLNITE
Mflv_1031|M.gilvum_PYR-GCK ------------DVCGKGLDAAVLTG------------------------
Mvan_5805|M.vanbaalenii_PYR-1 ------------DVCGKGLEAAVLTG------------------------
TH_3243|M.thermoresistible__bu ------------DVCGKGVEAAALTG------------------------
MAV_2108|M.avium_104 LQHAILGPADLPNGFAVRYQAATRPLQVGGDWFDIVDLEDGRVALVVGDC
MMAR_5186|M.marinum_M -------PVKPPPGFAVRYEPAVPPLEIGGDWYDVLPIGEHRIGIVVGDC
MSMEG_6131|M.smegmatis_MC2_155 ------------DVAGHGPDEAALGV------------------------
. :
Mb1399c|M.bovis_AF2122/97 DATINAARTVRAQLREWLAEIGADHSDIADIVHAISEFVENAVEHGYATD
Rv1364c|M.tuberculosis_H37Rv DATINAARTVRAQLREWLAEIGADHSDIADIVHAISEFVENAVEHGYATD
MUL_3853|M.ulcerans_Agy99 DATIHAARAIRSRLRQWLSEIGADSGDISDIVHAISEFVENAVEHGYSTD
Mflv_1031|M.gilvum_PYR-GCK ------------KIRNTLQALAPLAADHTGVLKLLNSALLSIDDTRFATL
Mvan_5805|M.vanbaalenii_PYR-1 ------------KIRNTLMALSPLAQDHAGVLRLLNAALLCVDDSRFATL
TH_3243|M.thermoresistible__bu ------------QTRQSLRTAAHFDRSPDVVLGAVNSVLCDLRVKQFVTV
MAV_2108|M.avium_104 VGHGLASATVMGQVRSACRALLIENPSPAAALAGLDRFAARLPGAQCTTA
MMAR_5186|M.marinum_M VGRGLPAAAIMGQLRSSARALLLTGAQPALLLEQLDAAASLIPDAYCTTV
MSMEG_6131|M.smegmatis_MC2_155 ------------ALRIGWRALTFAGLRGNERMRQLDRILTTERPGKGIFA
* : :.
Mb1399c|M.bovis_AF2122/97 VSKGIVVEA------------ALAGDGNVRASVIDRGQ------------
Rv1364c|M.tuberculosis_H37Rv VSKGIVVAA------------ALAGDGNVRASVIDRGQ------------
MUL_3853|M.ulcerans_Agy99 VSDGLAVAA------------ALGGDGKLRVAVIDHGT------------
Mflv_1031|M.gilvum_PYR-GCK VLASVVRR-----------------DGFVRLRLTSAGH------------
Mvan_5805|M.vanbaalenii_PYR-1 VLASVSRR-----------------DGQVLLRLTSAGH------------
TH_3243|M.thermoresistible__bu LCARIRARP----------------GGGVGVDLATAGH------------
MAV_2108|M.avium_104 VCAVLTPDTGELVYSSAGHPPPIVVHGDGSTQILDEGHTIALGIRRDWPR
MMAR_5186|M.marinum_M FLAILDTESGILDYSNAGHMPAVLAEPKSGPVLLTDARSVPLAVRRVEPR
MSMEG_6131|M.smegmatis_MC2_155 TLCSVALEP-----------------DSGRFTVVRAGH------------
: : .
Mb1399c|M.bovis_AF2122/97 ----------------WKDHRDGARGRGRGLAMAEALVSEARIMHGAGGT
Rv1364c|M.tuberculosis_H37Rv ----------------WKDHRDGARGRGRGLAMAEALVSEARIMHGAGGT
MUL_3853|M.ulcerans_Agy99 ----------------WKHHREGERGRGRGLAMAEALVSETHVSHDGAGT
Mflv_1031|M.gilvum_PYR-GCK ----------------LAPLIVRNDGS----------VEEADTGGTLVGV
Mvan_5805|M.vanbaalenii_PYR-1 ----------------LAPLIVRNDGR----------VEEAETAGTLVGV
TH_3243|M.thermoresistible__bu ----------------PAPLIVRGDGR----------VEQAEVYGTAAGL
MAV_2108|M.avium_104 PEARVTIPARATLLLYTDGLVERRRSPLDRGIYRVATVAQDARGSSLDEL
MMAR_5186|M.marinum_M PEATQVLPPGSTLLLFTDGLVERRYESIDEGLGRVADVVADSVMLPIDAV
MSMEG_6131|M.smegmatis_MC2_155 -----------------PGLLVHGNGT-------------VDWLEPRPGA
Mb1399c|M.bovis_AF2122/97 TATLTHRLSRPARFVTDTMVRRAAFQQ-TIDSEFVS-L---VESGRIVVR
Rv1364c|M.tuberculosis_H37Rv TATLTHRLSRPARFVTDTMVRRAAFQQ-TIDSEFVS-L---VESGRIVVR
MUL_3853|M.ulcerans_Agy99 TATVVHRLSRPAQFVTDPVVSRAPNRP-AIDSEFASTV---TESGHIVVR
Mflv_1031|M.gilvum_PYR-GCK MPTIRARTVDAALAPGETCL--------------------------LFTD
Mvan_5805|M.vanbaalenii_PYR-1 MPQIRARTVETSLAPGETCL--------------------------LYTD
TH_3243|M.thermoresistible__bu VSGMRYGATALELHRGDTMV--------------------------MFTD
MAV_2108|M.avium_104 ATRIMSAVAPSGGYQDDVVLLLYRHPA-PLELNFPADVNQLAPARTALRN
MMAR_5186|M.marinum_M ADAVLSQLAPTGGYDDDVAMVAYRYQHGPLRIETSATPDQLVIVRRQLLS
MSMEG_6131|M.smegmatis_MC2_155 ALGLGAREWPVNHYELPEGHG-----------------------LLLLTD
:
Mb1399c|M.bovis_AF2122/97 GDVDSTTAATLDRQIAVESRSGIAPVTIDLSAVTHLGSAGVGALAAACDR
Rv1364c|M.tuberculosis_H37Rv GDVDSTTAATLDRQIAVESRSGIAPVTIDLSAVTHLGSAGVGALAAACDR
MUL_3853|M.ulcerans_Agy99 GDVDSNTAPTLDRQIAVESRSGIASLTIDLSAVTHLGSAGVSALVTARDR
Mflv_1031|M.gilvum_PYR-GCK GVTEARGGPLGADMFGEARLSAALSQCAGLPAEAVVERIMMLTSEWVHNR
Mvan_5805|M.vanbaalenii_PYR-1 GVTEARGGPLGTDMFGEQRLSAALAECAGLPAEAVVERIMMLTTGWLNER
TH_3243|M.thermoresistible__bu GIDEAWG---AGGQYGMARLHALLPAYAGAAPEVVCEAVEQDVMEYLDGR
MAV_2108|M.avium_104 WLRRVRLDPEQTLNVLVAAGEAVANAIEHGHRHSPQGTIRLGATALGDQV
MMAR_5186|M.marinum_M WLQAAEVPDEQAGDIVLVVSEACTNCIEHAYRGQSVGTMLLDVEAADWAI
MSMEG_6131|M.smegmatis_MC2_155 GLFEGHAG-RGDERLGESGLLAVARALAATPGRDFVTSLINEAESRAQTH
. .
Mb1399c|M.bovis_AF2122/97 ARKQGTECVLVAPPGSPAHHVLSLVQLPVVGADTEDIFAQE---------
Rv1364c|M.tuberculosis_H37Rv ARKQGTECVLVAPPGSPAHHVLSLVQLPVVGADTEDIFAQE---------
MUL_3853|M.ulcerans_Agy99 ARKQGGRCVLMAPPGSPAHHVLSLVQIPVISGETENVFAED---------
Mflv_1031|M.gilvum_PYR-GCK AHDDIAVVAITAPRR---------THLSAVDGHTAGRFTA----------
Mvan_5805|M.vanbaalenii_PYR-1 AHDDIAVVAITAPRR---------THLSAVDGRTAGRYTA----------
TH_3243|M.thermoresistible__bu PHDDMAMVAVTCVR------------------------------------
MAV_2108|M.avium_104 RLTITDTGTWKVPQATSYPHRGRGIPLMKSLMNDVDIRSDTGGTTVQLSA
MMAR_5186|M.marinum_M HARVVDSGSWKKPAADP-GNGGRGMTLIRAISDEVEIDNSPGGTAIDMTF
MSMEG_6131|M.smegmatis_MC2_155 GGLSDDIAVIRVERSRR---------------------------------
Mb1399c|M.bovis_AF2122/97 ------
Rv1364c|M.tuberculosis_H37Rv ------
MUL_3853|M.ulcerans_Agy99 ------
Mflv_1031|M.gilvum_PYR-GCK ------
Mvan_5805|M.vanbaalenii_PYR-1 ------
TH_3243|M.thermoresistible__bu ------
MAV_2108|M.avium_104 RIT---
MMAR_5186|M.marinum_M RLPSED
MSMEG_6131|M.smegmatis_MC2_155 ------