For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RTLHVGDDGWPLMGEVIGRGELIIMLHGGGPDHHSMRPLADRLTGRYRVALPDIRGYGASRCPDPTLHRW DQYVTDTLAIMHALDATSAHLVGAGLGGTIALRTCLQHPQIARSAVIISAEAIEDDEDKAADAELMGRFA DRARSRGLQAAWELFVPYLQPLIANLVTEAIPRADAQSAAAAAAIGHDRAFATIEELRRVNAPTLVIAGN DPRHPTQLARTLADTLPHGVLAKATMSDLLVDADDIAQAFGPAIEQFLATTSDPDA*
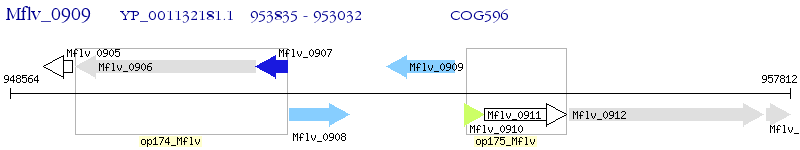
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0909 | - | - | 100% (267) | alpha/beta hydrolase fold |
| M. gilvum PYR-GCK | Mflv_2846 | - | e-134 | 87.59% (266) | alpha/beta hydrolase fold |
| M. gilvum PYR-GCK | Mflv_1348 | - | 9e-13 | 27.27% (264) | 3-oxoadipate enol-lactonase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2734 | - | 2e-09 | 28.12% (128) | hydrolase |
| M. tuberculosis H37Rv | Rv2715 | - | 2e-09 | 28.12% (128) | hydrolase |
| M. leprae Br4923 | MLBr_02297 | - | 1e-08 | 36.36% (99) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_0422c | - | 7e-09 | 36.36% (99) | epoxide hydrolase EphE |
| M. marinum M | MMAR_1502 | - | 2e-10 | 31.93% (119) | hypothetical protein MMAR_1502 |
| M. avium 104 | MAV_2517 | - | 2e-10 | 25.68% (292) | 2-hydroxy-6-ketonona-2,4-dienedioic acid hydrolase |
| M. smegmatis MC2 155 | MSMEG_1897 | pcaD | 7e-12 | 26.19% (252) | 3-oxoadipate enol-lactonase |
| M. thermoresistible (build 8) | TH_0963 | ephE | 1e-09 | 36.63% (101) | POSSIBLE EPOXIDE HYDROLASE EPHE (EPOXIDE HYDRATASE) |
| M. ulcerans Agy99 | MUL_4244 | ephE | 2e-09 | 38.38% (99) | epoxide hydrolase EphE |
| M. vanbaalenii PYR-1 | Mvan_2912 | - | 2e-09 | 33.33% (105) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2734|M.bovis_AF2122/97 ------MTERKRNLRPVRDVAPPTLQFRTVHGYRRAFRIAGSGP------
Rv2715|M.tuberculosis_H37Rv ------MTERKRNLRPVRDVAPPTLQFRTVHGYRRAFRIAGSGP------
Mvan_2912|M.vanbaalenii_PYR-1 ------MHD--------------DVKYLDLHGDRVAYREAGQGP------
MMAR_1502|M.marinum_M ------MTTTT-----------LAERTVTVDGKPIFFAEAGSGP------
MAV_2517|M.avium_104 ------MTTSN-----------MADHAVTVNGKSIFFAEKGTGA------
MLBr_02297|M.leprae_Br4923 ------MAMSNPSVTRIAG----PWHHLDVHANGIRFHVVEAVPTQPAGQ
MUL_4244|M.ulcerans_Agy99 ------MPAPDPSITRIDG----PWRHLDIHANGIRFHVVEAVP---AGQ
TH_0963|M.thermoresistible__bu ------MPPPDPSVTRIDG----PWRHHEVHANGIRFHVVEAEP---DEA
MAB_0422c|M.abscessus_ATCC_199 MLFDQTARHPDPSVVRISG----PWRHFDVHANGIRFHVVEADA----QD
MSMEG_1897|M.smegmatis_MC2_155 ---------------------------MSSVAVHHVVTGRSDGP------
Mflv_0909|M.gilvum_PYR-GCK ----------------------MRTLHVGDDGWPLMGEVIGRGE------
.
Mb2734|M.bovis_AF2122/97 ----------AILLIHGIGDNS---TTWNGVHA--KLAQRFTVIAPDLLG
Rv2715|M.tuberculosis_H37Rv ----------AILLIHGIGDNS---TTWNGVHA--KLAQRFTVIAPDLLG
Mvan_2912|M.vanbaalenii_PYR-1 ----------AVLLIHGMGGSS---LTWKALLP--HLATRYRVIAPDLLG
MMAR_1502|M.marinum_M ----------TVVLLHGGGPGATGLSNYARNID--LLAQRFRLIIPDMPG
MAV_2517|M.avium_104 ----------PVLLLHGGGPGASGVSNYSRNID--PLAEKFRVIVPDMPG
MLBr_02297|M.leprae_Br4923 DCAPSATARPLVMLLHGFGSFW---WSWRHQLR---GLTEARLVAVDLRG
MUL_4244|M.ulcerans_Agy99 RDNAPSTTQPLVMLLHGFGSFW---WSWRHQLR---GLTGARVVAVDLRG
TH_0963|M.thermoresistible__bu DRRIPRTERPLVILLHGFGSFW---YSWRHQLTGLTGLTGARVVAVDLRG
MAB_0422c|M.abscessus_ATCC_199 APTAPVTSRPLVLLLHGFGSFW---WSWRHQLG---ALPGARVVAVDLRG
MSMEG_1897|M.smegmatis_MC2_155 ----------AVVFSNSLGSTH---RMWDPQLA--AFEERFRVVRYDTRG
Mflv_0909|M.gilvum_PYR-GCK ----------LIIMLHGGGPDH---HSMRPLAD--RLTGRYRVALPDIRG
::: :. * : * *
Mb2734|M.bovis_AF2122/97 HGQSDK-PRADYSVAAYANGMRDLLSVLDIERVTIVGHSLGGGVAMQFAY
Rv2715|M.tuberculosis_H37Rv HGQSDK-PRADYSVAAYANGMRDLLSVLDIERVTIVGHSLGGGVAMQFAY
Mvan_2912|M.vanbaalenii_PYR-1 HGQSDK-PRGDYSLGAFAVWLRDLLDLLGIARVTLVGHSLGGGVAMQFVH
MMAR_1502|M.marinum_M YGRSSKGVDQSDPFGYLASAIRGLLDELRIPSAHLVGNSYGGAAALRLAL
MAV_2517|M.avium_104 YGRSAKGVDQSDPFGYLADTMRGLLDELGIDSAHLVGNSYGGSCALRLAL
MLBr_02297|M.leprae_Br4923 YGGSDK-PPRGYDGWTLAGDTAGLIRALGHSSATLVGHAEGGLACWATAL
MUL_4244|M.ulcerans_Agy99 YGGSDK-PPRGYDGWTLAGDTAGLIRALGHSSATLVGHADGGLACWTTAL
TH_0963|M.thermoresistible__bu YGGSDK-PPRGYDGWTLAGDTAGLVRALGHPRAALVGHADGGLVCWATAV
MAB_0422c|M.abscessus_ATCC_199 YGGSDK-PPRGYDGWTLAGDTAGLIRALGHTSATLVGHAEGGLVCWATAN
MSMEG_1897|M.smegmatis_MC2_155 HGQSPV-PEGPYTIDDLADDVVALLDTLGIERAHVVGLSLGGMTAMRLSA
Mflv_0909|M.gilvum_PYR-GCK YGASRCPDPTLHRWDQYVTDTLAIMHALDATSAHLVGAGLGGTIALRTCL
:* * . :: * . :** . ** .
Mb2734|M.bovis_AF2122/97 QFPQLVDRLILVSAGGVTKDVNIVFRLASLPMGSEAMALLRLPLVLPA--
Rv2715|M.tuberculosis_H37Rv QFPQLVDRLILVSAGGVTKDVNIVFRLASLPMGSEAMALLRLPLVLPA--
Mvan_2912|M.vanbaalenii_PYR-1 QHPDYCERLVLISSGGLGPELGRSLRLLSTPG----IELLLPVAAAPS--
MMAR_1502|M.marinum_M DSPHRVDRLVLMGPGGIGTTR-------SAPTAGLKTLLSYYAGDGPS--
MAV_2517|M.avium_104 DNPRRVGKLVLMGPGGIGTTR-------GLPTAGLNSLLAYYGGRGPS--
MLBr_02297|M.leprae_Br4923 LHPRLVRAIALINSPHPAALRRSMLTHRNQGYALLPTLLRYQLPIWPERL
MUL_4244|M.ulcerans_Agy99 LHPRLVRAIALISSPHPAALRRSTLTRRDQGRALLPTLLRYQVPLWPERL
TH_0963|M.thermoresistible__bu LHPRVVRSIALVSSPHPAALRASALSNRAQARALLPSLLSYQLPRWPERM
MAB_0422c|M.abscessus_ATCC_199 LHPRLVNAIAVISSPHPIALRTSTFRGTGQGGALLPSLMRYQVPILPERT
MSMEG_1897|M.smegmatis_MC2_155 RNPQRVDRIALLCTATHLPPS--------SAWTERAAAVRTHG-------
Mflv_0909|M.gilvum_PYR-GCK QHPQIARSAVIISAEAIEDDE---------DKAADAELMGRFADRARS--
* :: . :
Mb2734|M.bovis_AF2122/97 -----VQIAGRIVGKAIGTTSLG-HDLPNVLR-ILDDLPEPTASAAFGRT
Rv2715|M.tuberculosis_H37Rv -----VQIAGRIVGKAIGTTSLG-HDLPNVLR-ILDDLPEPTASAAFGRT
Mvan_2912|M.vanbaalenii_PYR-1 -----VVAVGERVRSWLGARGLASPEVGETWN-AYASLSDPQTRTAFLRT
MMAR_1502|M.marinum_M -----RAKLAHLIRTYLVYEGDSVPDELIDLR-YQASIDPAVIADPPLRR
MAV_2517|M.avium_104 -----RDKLEAFIRNYLVYDGASVPDELIDLR-YQASVDPEVVADPPLRR
MLBr_02297|M.leprae_Br4923 LTRNNAAEIERLVRSRSSAKWQACEDFAVTIDHLRIAIQIPAAAHCALEY
MUL_4244|M.ulcerans_Agy99 LTRDNAEEIERLIRSRTSAKWAASEDFSQSIGYLRRAIQIPAAAHCALEY
TH_0963|M.thermoresistible__bu LTKDDGARLERLVRSRAGAQWPATQDFATTVGHLRRAIQIPSAAHSALEY
MAB_0422c|M.abscessus_ATCC_199 LTRHRAAELERLVRSRAGTRWTSTDDFAQSMKHLRTAICIPGVAHCALEY
MSMEG_1897|M.smegmatis_MC2_155 -----SAAVAESVVGRWFTPGFLAADPGRKAAYQQMIAATPAEGYAG---
Mflv_0909|M.gilvum_PYR-GCK ------RGLQAAWELFVPYLQPLIANLVTEAIPRADAQSAAAAAAIG---
:
Mb2734|M.bovis_AF2122/97 LRA--VVDWRGQMVTMLDRCYLTEAIPVQIIWGTKDVVLPVRHAHMAHAA
Rv2715|M.tuberculosis_H37Rv LRA--VVDWRGQMVTMLDRCYLTEAIPVQIIWGTKDVVLPVRHAHMAHAA
Mvan_2912|M.vanbaalenii_PYR-1 LRS--VVDAQGQAVSALNRLHFTLGLPTLLIWGDQDRLIPPAHGEAAHAA
MMAR_1502|M.marinum_M PNG---LRTLWRMDLTRDRRLRQLPTPTLVLWGRDDKINRPAGGPKLLNL
MAV_2517|M.avium_104 PSGPMALRTLWRMDLARDRRLKRLRTPTLVLWGRDDKVNRPAGGPMLLNL
MLBr_02297|M.leprae_Br4923 QRWAVRSQLRNEGRKFMKSMSRPFSVPLLHVRGDADPYLLADSVDRTRRY
MUL_4244|M.ulcerans_Agy99 QRWAVRSQLRREGHRFMRSMNQRPGVPILHLRGDADPYVLAAPVERTQRY
TH_0963|M.thermoresistible__bu QRWAVRSQLRSEGHRFMRLMTRPVRVPVLHLHGESDPYVLADPVHRTQRH
MAB_0422c|M.abscessus_ATCC_199 QRWAVRSQLRSEGWRFMRSMNRELSVPVLHMRGESDPYVLADPVHRSLPY
MSMEG_1897|M.smegmatis_MC2_155 -------CCEAIAALDLRADLARITAPTLAIAGTDDPATPPDRLAEITAA
Mflv_0909|M.gilvum_PYR-GCK ----------HDRAFATIEELRRVNAPTLVIAG-NDPRHPTQLARTLADT
* : * *
Mb2734|M.bovis_AF2122/97 MPGSQLEIFEGSGHFPFHD-DPARFIDIVERFMDTTEPAEYDQAALRALL
Rv2715|M.tuberculosis_H37Rv MPGSQLEIFEGSGHFPFHD-DPARFIDIVERFMDTTEPAEYDQAALRALL
Mvan_2912|M.vanbaalenii_PYR-1 LPGSRLVILPAVGHFPQVE-APLAVADTLDDFIANSSP------------
MMAR_1502|M.marinum_M MPNAELVMTSRTGHWMQWE-RAELFNQLVAEFLSQSSAFAS---------
MAV_2517|M.avium_104 MPNAELVMTSRTGHWVQWE-RADLFNKLVTEFLHNTSVFAQ---------
MLBr_02297|M.leprae_Br4923 APHGRYVSVSDAGHFSHEE-APEEINRYLTRFLKQVHGPRLS--------
MUL_4244|M.ulcerans_Agy99 APHGRYVSVAGVGHYSHEE-APEEINRHLMRFFEQTRDARLS--------
TH_0963|M.thermoresistible__bu APHGRYVSVPGAGHFAHEE-APAVVNEELGRFLTGRTGR-----------
MAB_0422c|M.abscessus_ATCC_199 APNGQFVALRGVGHYVHEE-DPAAVNEHLRRFTGHR--------------
MSMEG_1897|M.smegmatis_MC2_155 VRGARLVQVTPGAHLANVE-QADAITPTLLEHLGTEQS------------
Mflv_0909|M.gilvum_PYR-GCK LPHGVLAKATMSDLLVDADDIAQAFGPAIEQFLATTSDPDA---------
. : . . : .
Mb2734|M.bovis_AF2122/97 RRGGGEATVTGSADTRVAVLNAIGSNERSAT
Rv2715|M.tuberculosis_H37Rv RRGGGEATVTGSADTRVAVLNAIGSNERSAT
Mvan_2912|M.vanbaalenii_PYR-1 WHGPPRHADHAEAD-----------------
MMAR_1502|M.marinum_M -------------------------------
MAV_2517|M.avium_104 -------------------------------
MLBr_02297|M.leprae_Br4923 -------------------------------
MUL_4244|M.ulcerans_Agy99 -------------------------------
TH_0963|M.thermoresistible__bu -------------------------------
MAB_0422c|M.abscessus_ATCC_199 -------------------------------
MSMEG_1897|M.smegmatis_MC2_155 -------------------------------
Mflv_0909|M.gilvum_PYR-GCK -------------------------------