For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPAPDPSITRIDGPWRHLDIHANGIRFHVVEAVPAGQRDNAPSTTQPLVMLLHGFGSFWWSWRHQLRGLT GARVVAVDLRGYGGSDKPPRGYDGWTLAGDTAGLIRALGHSSATLVGHADGGLACWTTALLHPRLVRAIA LISSPHPAALRRSTLTRRDQGRALLPTLLRYQVPLWPERLLTRDNAEEIERLIRSRTSAKWAASEDFSQS IGYLRRAIQIPAAAHCALEYQRWAVRSQLRREGHRFMRSMNQRPGVPILHLRGDADPYVLAAPVERTQRY APHGRYVSVAGVGHYSHEEAPEEINRHLMRFFEQTRDARLS
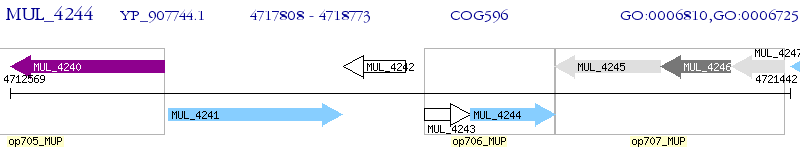
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4244 | ephE | - | 100% (321) | epoxide hydrolase EphE |
| M. ulcerans Agy99 | MUL_4193 | ephA | 1e-22 | 30.72% (319) | epoxide hydrolase EphA |
| M. ulcerans Agy99 | MUL_2888 | ephB | 3e-18 | 39.85% (133) | epoxide hydrolase EphB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3694 | ephE | 1e-159 | 82.61% (322) | epoxide hydrolase EphE |
| M. gilvum PYR-GCK | Mflv_1377 | - | 1e-144 | 75.08% (313) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3670 | ephE | 1e-159 | 82.61% (322) | epoxide hydrolase EphE |
| M. leprae Br4923 | MLBr_02297 | - | 1e-151 | 79.38% (325) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_0422c | - | 1e-132 | 71.43% (308) | epoxide hydrolase EphE |
| M. marinum M | MMAR_5158 | ephE | 0.0 | 99.38% (321) | epoxide hydrolase EphE |
| M. avium 104 | MAV_0458 | - | 1e-156 | 82.43% (313) | hydrolase, alpha/beta fold family protein |
| M. smegmatis MC2 155 | MSMEG_6184 | - | 1e-138 | 72.47% (316) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_0963 | ephE | 1e-138 | 72.29% (314) | POSSIBLE EPOXIDE HYDROLASE EPHE (EPOXIDE HYDRATASE) |
| M. vanbaalenii PYR-1 | Mvan_5429 | - | 1e-145 | 76.58% (316) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1377|M.gilvum_PYR-GCK ------MTPPDPSVVRIGGPWRHLDVHANGIRFHVVEAQRPSGADDVTR-
MSMEG_6184|M.smegmatis_MC2_155 ------MPPPDPSVTRIDGPWRHLQVHANGIRFHVVEAAPEAVEPGVT--
Mvan_5429|M.vanbaalenii_PYR-1 ------MVRPDPSVVRIGGPWRHLDVHANGIRFHVVEAERQAGADDRSK-
TH_0963|M.thermoresistible__bu ------MPPPDPSVTRIDGPWRHHEVHANGIRFHVVEAEPDEADRRIPR-
MUL_4244|M.ulcerans_Agy99 ------MPAPDPSITRIDGPWRHLDIHANGIRFHVVEAVPAGQRDN----
MMAR_5158|M.marinum_M ------MPAPDPSITRIDGPWRHLDIHANGIRFHVVEAIPAGQRDN----
Mb3694|M.bovis_AF2122/97 ------MAAPDPSMTRIAGPWRHLDVHANGIRFHVVEAVPSGQPEGPDAA
Rv3670|M.tuberculosis_H37Rv ------MAAPDPSMTRIAGPWRHLDVHANGIRFHVVEAVPSGQPEGPDAA
MLBr_02297|M.leprae_Br4923 ------MAMSNPSVTRIAGPWHHLDVHANGIRFHVVEAVPT-QPAG----
MAV_0458|M.avium_104 ------MAPPDPSVTRIDGPWRHLDVHANGIRFHVVEALPVGDARG----
MAB_0422c|M.abscessus_ATCC_199 MLFDQTARHPDPSVVRISGPWRHFDVHANGIRFHVVEADAQDAPTA----
.:**:.** ***:* ::************
Mflv_1377|M.gilvum_PYR-GCK -----PLTDRPLVILLHGFGSFWWSWRHQLTGLS---GARLVAVDLRGYG
MSMEG_6184|M.smegmatis_MC2_155 -------SDRPLVILLHGFGSFWWSWRHQLTGLR---DARVVAVDLRGYG
Mvan_5429|M.vanbaalenii_PYR-1 -----PLTDRPLVILLHGFGSFWWSWRHQLAGLT---GARVVAVDLRGYG
TH_0963|M.thermoresistible__bu -------TERPLVILLHGFGSFWYSWRHQLTGLTGLTGARVVAVDLRGYG
MUL_4244|M.ulcerans_Agy99 ----APSTTQPLVMLLHGFGSFWWSWRHQLRGLT---GARVVAVDLRGYG
MMAR_5158|M.marinum_M ----APSTTQPLVMLLHGFGSFWWSWRHQLRGLT---GARVVAVDLRGYG
Mb3694|M.bovis_AF2122/97 TPPMQPALARPLVILLHGFGSFWWSWRHQLCGLT---GARVVAVDLRGYG
Rv3670|M.tuberculosis_H37Rv TPPMQPALARPLVILLHGFGSFWWSWRHQLCGLT---GARVVAVDLRGYG
MLBr_02297|M.leprae_Br4923 QDCAPSATARPLVMLLHGFGSFWWSWRHQLRGLT---EARLVAVDLRGYG
MAV_0458|M.avium_104 ----LPATARPLVMLLHGFGSFWWSWRHQLRGLT---GARVVAVDLRGYG
MAB_0422c|M.abscessus_ATCC_199 -----PVTSRPLVLLLHGFGSFWWSWRHQLGALP---GARVVAVDLRGYG
:***:*********:****** .* **:*********
Mflv_1377|M.gilvum_PYR-GCK GSDKPPRGYDGWTLAGDTAGLVRALGHNSATLVGHADGGLVCWATSVLHP
MSMEG_6184|M.smegmatis_MC2_155 GSDKPPRGYDGWTLAGDTAGLVRALGHTSATLVGHADGGLVCWATSVLHP
Mvan_5429|M.vanbaalenii_PYR-1 GSDKPPRGYDGWTLAGDAAGLVRALGHNTATLVGHADGGLVCWATSVLHP
TH_0963|M.thermoresistible__bu GSDKPPRGYDGWTLAGDTAGLVRALGHPRAALVGHADGGLVCWATAVLHP
MUL_4244|M.ulcerans_Agy99 GSDKPPRGYDGWTLAGDTAGLIRALGHSSATLVGHADGGLACWTTALLHP
MMAR_5158|M.marinum_M GSDKPPRGYDGWTLAGDTAGLIRALGHSSATLVGHADGGLACWTTALLHP
Mb3694|M.bovis_AF2122/97 GSDKPPRGYDGWTLAGDTAGLIRALGHPSATLVGHADGGLACWTTALLHS
Rv3670|M.tuberculosis_H37Rv GSDKPPRGYDGWTLAGDTAGLIRALGHPSATLVGHADGGLACWTTALLHS
MLBr_02297|M.leprae_Br4923 GSDKPPRGYDGWTLAGDTAGLIRALGHSSATLVGHAEGGLACWATALLHP
MAV_0458|M.avium_104 GSDKPPRGYDGWTLSGDAAGLIRALGHSSATLVGHADGGLACWATALLHA
MAB_0422c|M.abscessus_ATCC_199 GSDKPPRGYDGWTLAGDTAGLIRALGHTSATLVGHAEGGLVCWATANLHP
**************:**:***:***** *:*****:***.**:*: **.
Mflv_1377|M.gilvum_PYR-GCK RVVKAIAVVSSPHPSALRASALRRRDQGRALLPSMMRYQVPMWPERALTR
MSMEG_6184|M.smegmatis_MC2_155 RAVNAIAVISSPHPVALRASALRRRDQGRALLPWMLGYQVPMWPERKLTR
Mvan_5429|M.vanbaalenii_PYR-1 RVVQAIAVVSSPHPAALRASALTRRDQGQALLPSLLRYQVPIRPERLLTR
TH_0963|M.thermoresistible__bu RVVRSIALVSSPHPAALRASALSNRAQARALLPSLLSYQLPRWPERMLTK
MUL_4244|M.ulcerans_Agy99 RLVRAIALISSPHPAALRRSTLTRRDQGRALLPTLLRYQVPLWPERLLTR
MMAR_5158|M.marinum_M RLVRAIALISSPHPAALRRSTLTRRDQGRALLPTLLRYQVPLWPERLLTR
Mb3694|M.bovis_AF2122/97 RLVRAIALISSPHPAALRRSTLTRRDQRHALLPTLLRYQLPIWPERLLTR
Rv3670|M.tuberculosis_H37Rv RLVRAIALISSPHPAALRRSTLTRRDQRHALLPTLLRYQLPIWPERLLTR
MLBr_02297|M.leprae_Br4923 RLVRAIALINSPHPAALRRSMLTHRNQGYALLPTLLRYQLPIWPERLLTR
MAV_0458|M.avium_104 RLVSAVALISSPHPAALRQSTLARRDQAGALLPTLLRYQVPLWPERLLTR
MAB_0422c|M.abscessus_ATCC_199 RLVNAIAVISSPHPIALRTSTFRGTGQGGALLPSLMRYQVPILPERTLTR
* * ::*::.**** *** * : * **** :: **:* *** **:
Mflv_1377|M.gilvum_PYR-GCK HNGAELERLVRSRASEKWQVSEDFAETVGHLRRAVQIPSAAHCALEYQRW
MSMEG_6184|M.smegmatis_MC2_155 DDAAELERVVRSRAGDAWQQTPDFAETMRHLRRAIQIPGAAHSALEYQRW
Mvan_5429|M.vanbaalenii_PYR-1 DDGAELERLVRSRSSAKWQAGEDFSESIRHMRRAIQIPSAAHCALEYQRW
TH_0963|M.thermoresistible__bu DDGARLERLVRSRAGAQWPATQDFATTVGHLRRAIQIPSAAHSALEYQRW
MUL_4244|M.ulcerans_Agy99 DNAEEIERLIRSRTSAKWAASEDFSQSIGYLRRAIQIPAAAHCALEYQRW
MMAR_5158|M.marinum_M DNAEEIERLIRSRTSAKWAASEDFSQSIGYLRRAIQIPAAAHCALEYQRW
Mb3694|M.bovis_AF2122/97 NNAAEIERLVRARGCAKWLASEDFSQAIDHLRQAIQIPAAAHCALEYQRW
Rv3670|M.tuberculosis_H37Rv NNAAEIERLVRARGCAKWLASEDFSQAIDHLRQAIQIPAAAHCALEYQRW
MLBr_02297|M.leprae_Br4923 NNAAEIERLVRSRSSAKWQACEDFAVTIDHLRIAIQIPAAAHCALEYQRW
MAV_0458|M.avium_104 DGGAEIERLVRSRSCAKWLASEDFSETIGHLRTAIQIPAAAHCALEYQRW
MAB_0422c|M.abscessus_ATCC_199 HRAAELERLVRSRAGTRWTSTDDFAQSMKHLRTAICIPGVAHCALEYQRW
. . .:**::*:* * **: :: ::* *: **..**.*******
Mflv_1377|M.gilvum_PYR-GCK AVRSQLRAEGRRFMRSMRAPLSVPMLHLRGDADPYVLPDPVYRTQRYAPH
MSMEG_6184|M.smegmatis_MC2_155 AVRSQLRGEGRRFMASMRRPVNVPVLHLRGDEDPYVLPDPVYRTQRYAPH
Mvan_5429|M.vanbaalenii_PYR-1 AVRSQLRAEGRRFMSSMKRPLGVPVLHLRGEQDPYVLADPVYRTQRYAPH
TH_0963|M.thermoresistible__bu AVRSQLRSEGHRFMRLMTRPVRVPVLHLHGESDPYVLADPVHRTQRHAPH
MUL_4244|M.ulcerans_Agy99 AVRSQLRREGHRFMRSMNQRPGVPILHLRGDADPYVLAAPVERTQRYAPH
MMAR_5158|M.marinum_M AVRSQLRGEGHRFMRSMNQRPGVPILHLRGDADPYVLAAPVERTQRYAPH
Mb3694|M.bovis_AF2122/97 AVRSQLRSEGRRFIRAMTQQLGMPLLHLRGDADPYVLADPVERTQRYAPH
Rv3670|M.tuberculosis_H37Rv AVRSQLRSEGRRFIRAMTQQLGMPLLHLRGDADPYVLADPVERTQRYAPH
MLBr_02297|M.leprae_Br4923 AVRSQLRNEGRKFMKSMSRPFSVPLLHVRGDADPYLLADSVDRTRRYAPH
MAV_0458|M.avium_104 AVRSQLRDEGRRFLRSMRRQLGVPLLHLRGEEDPYVLADPVDRTRRYAPQ
MAB_0422c|M.abscessus_ATCC_199 AVRSQLRSEGWRFMRSMNRELSVPVLHMRGESDPYVLADPVHRSLPYAPN
******* ** :*: * :*:**::*: ***:*. .* *: :**:
Mflv_1377|M.gilvum_PYR-GCK GRYVSIAGAGHFAHEEQPAAVNAQLSRFLSQVYT----
MSMEG_6184|M.smegmatis_MC2_155 GRYVSITGAGHFAHEEQPDAVNEQLGRFLAQIRS----
Mvan_5429|M.vanbaalenii_PYR-1 GRYVSIEGAGHFSHEEAPAAVNEQLRRFLNQVYG----
TH_0963|M.thermoresistible__bu GRYVSVPGAGHFAHEEAPAVVNEELGRFLTGRTGR---
MUL_4244|M.ulcerans_Agy99 GRYVSVAGVGHYSHEEAPEEINRHLMRFFEQTRDARLS
MMAR_5158|M.marinum_M GRYVSVAGVGHYSHEEAPEEINRHLMRFFEQTRDARLS
Mb3694|M.bovis_AF2122/97 GRYISIAGAGHFSHEEAPEEVNRHLMRFLEQVHQLS--
Rv3670|M.tuberculosis_H37Rv GRYISIAGAGHFSHEEAPEEVNRHLMRFLEQVHQLS--
MLBr_02297|M.leprae_Br4923 GRYVSVSDAGHFSHEEAPEEINRYLTRFLKQVHGPRLS
MAV_0458|M.avium_104 GRYVTVAGAGHFSHEEAPEEINRHLTSFLEDVHGPAVN
MAB_0422c|M.abscessus_ATCC_199 GQFVALRGVGHYVHEEDPAAVNEHLRRFTGHR------
*::::: ..**: *** * :* * *