For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTDLRGRVALVTGAAQGMGAAHARRLAKAGATVAVNDREDSDALAALASEIGGLTAAGDVSDPAQCRAVT ARIAETAGRLDILVANHAYMTMAPLLEHDPDDWWKVVDTNLGGTFFLVQSVLPYMREAGAGRIVVISSEW GVTGWPGATAYAASKSGLISLVKTLGRELAREHIIVNAVAPGVIDTPQLQVDADAAGVALEVIHRRYAES IPLGRIGVGDEVSAAVELLADFDVEAIVGQVISCNGGSMRARA*
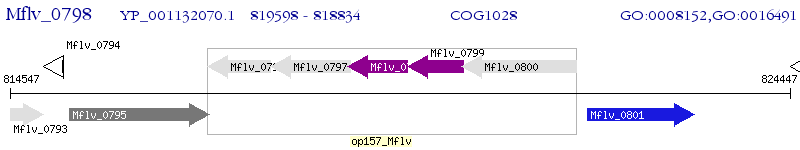
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0798 | - | - | 100% (254) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_2469 | - | 2e-40 | 39.92% (253) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_5072 | fabG | 5e-31 | 34.86% (284) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 4e-29 | 34.94% (249) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv2857c | - | 4e-29 | 34.94% (249) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 2e-29 | 35.95% (242) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0952 | - | 2e-30 | 34.65% (254) | short chain dehydrogenase |
| M. marinum M | MMAR_0387 | fabG3 | 7e-33 | 37.30% (244) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. avium 104 | MAV_3962 | - | 6e-37 | 38.71% (248) | NAD-dependent epimerase/dehydratase:Short-chain |
| M. smegmatis MC2 155 | MSMEG_1071 | - | 1e-109 | 78.57% (252) | 3-oxoacyl- |
| M. thermoresistible (build 8) | TH_3954 | - | 2e-30 | 37.30% (244) | PUTATIVE 3-oxoacyl-[acyl-carrier-protein] reductase |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 6e-33 | 37.30% (244) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. vanbaalenii PYR-1 | Mvan_0045 | - | 1e-122 | 85.43% (254) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0798|M.gilvum_PYR-GCK --------------MTTDLRGRVALVTGAAQGMGAAHARRLAKAGATVAV
Mvan_0045|M.vanbaalenii_PYR-1 --------------MSTDLRGRVALVTGAARGMGAAHARRLAAAGATVAV
MSMEG_1071|M.smegmatis_MC2_155 ---------------MTSLAGRVALVTGAAQGMGAAHARRLASAGATVAV
Mb2882c|M.bovis_AF2122/97 -----------MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVV
Rv2857c|M.tuberculosis_H37Rv -----------MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVV
MMAR_0387|M.marinum_M ---------------MGRVDGKVALISGGARGMGASHARLLVQEGAKVVI
MUL_1037|M.ulcerans_Agy99 ---------------MGRVDGKVALISGGARGMGASHARLLVQEGAKVVI
TH_3954|M.thermoresistible__bu -------------MPSALLAEKVAVVTGAGRGIGREIARVLHAHGAEVVL
MAV_3962|M.avium_104 --------------------MRVAVVTGGASGMGEATCHELGRRGMKIAV
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
MAB_0952|M.abscessus_ATCC_1997 --MVVESAEPAERKVMFDLSARSVLVTGGTKGIGRGIATVFARAGANVAV
: :::*. *:* : * :.:
Mflv_0798|M.gilvum_PYR-GCK NDREDS-------DALAALASEIG----GLTAAGDVSDPAQCRAVTARIA
Mvan_0045|M.vanbaalenii_PYR-1 NDREDS-------DALTALATEIG----GLTAPGDVSDPKSCRALATHVA
MSMEG_1071|M.smegmatis_MC2_155 NDIRAG-------DALHALAQEIG----GLTAPGDVSDPAVCRQIADDVA
Mb2882c|M.bovis_AF2122/97 GDVDV--------EAGGAAADEL----SGLFVPTDVCDEDAVNGLFDGAA
Rv2857c|M.tuberculosis_H37Rv GDVDV--------EAGGAAADEL----SGLFVPTDVCDEDAVNGLFDGAA
MMAR_0387|M.marinum_M GDILD--------EEGKALAEEIGD--AARYVHLDVTQPDQWEAAVATAV
MUL_1037|M.ulcerans_Agy99 GDILD--------EEGKALAEEIGD--AARYVHLDVTQPDQWEAAVATAV
TH_3954|M.thermoresistible__bu ADLDA--------DAAAQAAAEVDG--TG--VGCDVTAEDQMKRLVADTV
MAV_3962|M.avium_104 LDVDEH-------AAQRVTDDLRTGGATALGVGADVTDRAAVEQAFAKVR
MLBr_01807|M.leprae_Br4923 THRAS------------VVPDWFFG------VECDVTDNDAIGAAFKAVE
MAB_0952|M.abscessus_ATCC_1997 AARSPRELSSVTAELGELGAGNVIG------VRLDVSDPGSCADAARTVV
. **
Mflv_0798|M.gilvum_PYR-GCK ETAGRLDILVANHAYMT--MAPLLEHDPDDWWKVVDTNLGGTFFLVQSVL
Mvan_0045|M.vanbaalenii_PYR-1 ATTGRLDVLVANHAYMT--MAPLLEHADDDWWKVVDTNLGGTFFLVQSVL
MSMEG_1071|M.smegmatis_MC2_155 AATGRLDILVANHAYMT--MAPLLDHDEGDWWRVVDTNLGGTFHLVQAVL
Mb2882c|M.bovis_AF2122/97 ETYGRIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAAL
Rv2857c|M.tuberculosis_H37Rv ETYGRIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAAL
MMAR_0387|M.marinum_M DEFGKLDVLVNNAGIVA--LGQLKKFDLGKWQKVIDVNLTGTFLGMRAAV
MUL_1037|M.ulcerans_Agy99 DEFGKLDVLVNNVGIVA--LGQLKKFDLGKWQKVIDVNLTGTFLGMRAAV
TH_3954|M.thermoresistible__bu AKHGRLDLFVNNAGITR--DASLKRMTVADFDAVVTVHLRGTWLGVREAT
MAV_3962|M.avium_104 SELGPVTVLVTSAGMFG--FSPFLDITAESWSRIIDVNLTGTFHCCQVAL
MLBr_01807|M.leprae_Br4923 EHQGPVEVLVANAGITS--DMLIMRMSEEQFQKVIDVNLTGVFRVAKLAS
MAB_0952|M.abscessus_ATCC_1997 DAFGALDVVCANAGIFP--EARLDTMTPEQLSEVLDVNVKGTVYTVQACL
* : : . . : : .:: .. :
Mflv_0798|M.gilvum_PYR-GCK PYMREAGAGRIVVISSEWGVTG-WPGATAYAASKSGLISLVKTLGRELAR
Mvan_0045|M.vanbaalenii_PYR-1 PHMRRTGAGRIVVIASEWGVTG-WPEATAYAAAKSGLISLVKTLGRELAR
MSMEG_1071|M.smegmatis_MC2_155 PHMRRLGAGRIVVIASEWGVIG-WPEATAYAAAKSGLISLVKSLGRELAP
Mb2882c|M.bovis_AF2122/97 RHMVLAGKGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFAR
Rv2857c|M.tuberculosis_H37Rv RHMVLAGKGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFAR
MMAR_0387|M.marinum_M EPMTAAGSGSIINVSSIEGLRG-APAVHPYVASKWAVRGLTKSAALELAP
MUL_1037|M.ulcerans_Agy99 EPMTAAGSGSIINVSSIEGLRG-APAVHPYVASKWAVRGLTKSAALELAP
TH_3954|M.thermoresistible__bu AVMREQKSGSIVNISSLSGKSG-NPGQTNYSAAKAGIIGLTKAAAKEVAH
MAV_3962|M.avium_104 PDMVAANWGRIVMISSSSAQRG-SPFAAHYAASKGAVITLTKSLAREYAP
MLBr_01807|M.leprae_Br4923 RNMIKQRFGRYIFMGSVVG-LSGVPGQANYAASKSGLIGMARSLARELGS
MAB_0952|M.abscessus_ATCC_1997 APLTASGRGRVILTSSITGPVTGYPGWSHYGASKAAQLGFMRTAAIELAP
: * : .* . . * *:* . : : . : .
Mflv_0798|M.gilvum_PYR-GCK EHIIVNAVAPGVIDTPQLQVDADAAGVALEVIHRRYAESIPLGRIGVGDE
Mvan_0045|M.vanbaalenii_PYR-1 EHIIVNAVAPGVIDTPQLQVDADAAGVGLDDIHRRYAEAIPLGRIGSADE
MSMEG_1071|M.smegmatis_MC2_155 EHIIVNAVAPGVTDTPQLQVDADAAGVDLATIHAQYAEAIPMGRIGSADE
Mb2882c|M.bovis_AF2122/97 QGIRVNALCPGPVNTPLLQELFAKN----PERAARRMVHVPLGRFAEPDE
Rv2857c|M.tuberculosis_H37Rv QGIRVNALCPGPVNTPLLQELFAKN----PERAARRMVHVPLGRFAEPDE
MMAR_0387|M.marinum_M LNIRVNSIHPGFIRTPMTANLP------------DDMVTIPLGRPAESRE
MUL_1037|M.ulcerans_Agy99 LNIRVNSIHPGFIRTPMTANLP------------DDMVTIPLGRPAESRE
TH_3954|M.thermoresistible__bu HNVRVNAIQPGLIRTPMTETMPA-------EVFAEREAAVPMKRAGEPAE
MAV_3962|M.avium_104 HGITVNNIPPSGIETPMQHQGQAAG---YLPSNEQIAANIPLGYLGTGAD
MLBr_01807|M.leprae_Br4923 RSITANVVAPGFIDTEMTRAMDSKY-------QEAALQAIPLGRIGAVEE
MAB_0952|M.abscessus_ATCC_1997 RGVTVNAILPGNILTEGLVDMGEEY-------ISGMARSIPMGMLGSPVD
: .* : *. * :*: . :
Mflv_0798|M.gilvum_PYR-GCK VSAAVELLADFDVEAIVGQVISCNGGSMRARA------
Mvan_0045|M.vanbaalenii_PYR-1 VAVAVELLSDFDLEAVVGQVISCNGGSTRTRA------
MSMEG_1071|M.smegmatis_MC2_155 IAATVEFLADFELEAVVGQVISSNGGSTRARA------
Mb2882c|M.bovis_AF2122/97 IAAAVAFLASDDASFITASTFLVDGGISSAYVTPL---
Rv2857c|M.tuberculosis_H37Rv IAAAVAFLASDDASFITASTFLVDGGISSAYVTPL---
MMAR_0387|M.marinum_M VSTFVVFLASDDASYATGSEFVMDGGLVTDVPHKQF--
MUL_1037|M.ulcerans_Agy99 VSTFVVFLASDDASYATGSEFVMDGGLVTDVPHKQF--
TH_3954|M.thermoresistible__bu VAGAVVFLGSDLSSYITGSVIEVGGGRYM---------
MAV_3962|M.avium_104 IAAAVGFLTSDEARYITGQVLGVNGGAVMLTVRIR---
MLBr_01807|M.leprae_Br4923 IAGAVSFLASEDAGYISGAVIPVDGGLGMGH-------
MAB_0952|M.abscessus_ATCC_1997 IGHLAAFLATDEAGYITGQAIVVDGGQVLPESPDAVNP
:. . :* . : .**