For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGDWLAARRTEVAADRILDAAGELFAAQSAATVGMHEIAAAAGCSRATLYRYFENREALYTAYVHRESYR LYREMTEQIMSFDDPRERLIEGMLSSLRNVRESPALSSWFATTQRPIGAEMAEQSEVIKALTEAFVISLG PDDADARASGATGEGSVAHRSRWLVRVMTSLMLFPGHDEADERAMLEEFVVPIVLPRRQADQATQ*
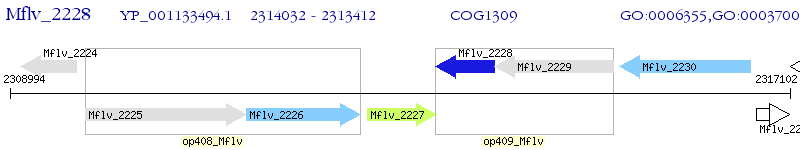
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2228 | - | - | 100% (206) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3474 | - | 1e-14 | 29.32% (191) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4624 | - | 3e-14 | 28.88% (187) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1805c | - | 2e-12 | 25.65% (191) | transcriptional regulatory protein |
| M. tuberculosis H37Rv | Rv1255c | - | 1e-61 | 59.90% (202) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_02568 | - | 2e-06 | 30.61% (98) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1938c | - | 4e-17 | 28.35% (194) | TetR family transcriptional regulator |
| M. marinum M | MMAR_4185 | - | 2e-61 | 60.31% (194) | transcriptional regulatory protein |
| M. avium 104 | MAV_1403 | - | 5e-62 | 61.34% (194) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_5040 | - | 1e-69 | 66.83% (199) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2228 | - | 9e-63 | 62.56% (195) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_4487 | - | 1e-61 | 60.31% (194) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_4466 | - | 2e-93 | 86.07% (201) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1805c|M.bovis_AF2122/97 -MPG----NDWIVGGNRRTIAAERIYAAATDLITRYGLNALDIDKLAREV
MAB_1938c|M.abscessus_ATCC_199 MSPY----ADWLHGSDRHTIAADRIVSVAAKQIARHGLDRLNLDAVAREA
Mflv_2228|M.gilvum_PYR-GCK -MAG-----DWLAA-RRTEVAADRILDAAGELFAAQSAATVGMHEIAAAA
Mvan_4466|M.vanbaalenii_PYR-1 -MAG-----DWLAA-RRTEVAADRILDAAGELFAAQPAATVGMHEIASAA
MSMEG_5040|M.smegmatis_MC2_155 -MA------DWLAG-RRGEVAADLILDAADALFARKDAATVGMHEIAAAA
TH_2228|M.thermoresistible__bu -MTG-----NWLAD-RRTEVAAERILDAAHELFTRHDAATVGMRDIARAA
MMAR_4185|M.marinum_M -MG----RTDWLAA-HRSEVAADRILDAAERLYTEHDPASIGMNEIARAA
MUL_4487|M.ulcerans_Agy99 -MG----RTDWLAA-HRSEVAADRILDAAERLYTEHDPASIGMNEIARAA
MAV_1403|M.avium_104 -MPGARRSTDWLSG-HRNEAAVDRILDAAQELYTQRDSESIGMNEIARAA
Rv1255c|M.tuberculosis_H37Rv -MAG----TDWLSA-RRTELAADRILDAAERLFTQRDPASIGMNEIAKAA
MLBr_02568|M.leprae_Br4923 -MAG-------GAKRIPRAVREQQMLDAAVQMFSVNGYHKTSMDAIAAEA
: : .* : .: :* .
Mb1805c|M.bovis_AF2122/97 HCSRATIYRRAGGKAQIRDVVLTRAAARIADGVRSDVETLRG-RERVVAA
MAB_1938c|M.abscessus_ATCC_199 GCSRATVYRHVGGKTAIVDAVLARNIAKVTADIQRAVQSADA-RRRAVVA
Mflv_2228|M.gilvum_PYR-GCK GCSRATLYRYFENREALYTAYVHRESYRLYREMTEQIMSFDDPRERLIEG
Mvan_4466|M.vanbaalenii_PYR-1 GCSRATLYRYFENREALYTAYVHREAYRLYVEMTDQITSIADPRERLIEG
MSMEG_5040|M.smegmatis_MC2_155 GCSRATLYRYFENRDALYTAYVHREARRVYQEVSDQVVGIGDPAERLMEG
TH_2228|M.thermoresistible__bu GCSRATLYRYFDSREALHTAYVHREARRLYRELGPRLAAIGDPQNRLIEG
MMAR_4185|M.marinum_M GCSRATLYRYFESREALRTAYVHRETLRLGREIMQQIGDIDDPRDQLITS
MUL_4487|M.ulcerans_Agy99 GCSRATLYRYFESREALRTAYVHRETLRLGREIMQQIGDIDDPRDQLITS
MAV_1403|M.avium_104 GCSRATLYRYFENREALRTAYVHREANRLGRAIMEQIDGIDEPRQRLTAS
Rv1255c|M.tuberculosis_H37Rv GCSRATLYRYFDSREALRTAYVHRETRRLGREIMVKIADVVEPAERLLVS
MLBr_02568|M.leprae_Br4923 QISKPMLYLYYGSKEDLFGACLNREMSRFIAMVRADIDFTQSPKDLLRNT
*:. :* .: : . : * :. : :
Mb1805c|M.bovis_AF2122/97 ILLSLQRIRSD--PLGKLMFGSIHGGAGELAWLTESPLLADFATELTGIA
MAB_1938c|M.abscessus_ATCC_199 ITASLTAIRHD--PVISTMIATMSP-TSLGSYLNASSEIPRSASAFTGLA
Mflv_2228|M.gilvum_PYR-GCK MLSSLRNVRES--PALSSWFATTQR-PIGAEMAEQSEVIKALTEAFVISL
Mvan_4466|M.vanbaalenii_PYR-1 MLASLRNVRES--PALASWFATTQR-PIGAEMAEESEVIKALTEAFVISL
MSMEG_5040|M.smegmatis_MC2_155 VITALRAVRCS--PALSSWFAATQR-PIGGEMAEHSEVIRALVEGFVRSL
TH_2228|M.thermoresistible__bu ILAALDAVREN--PALSSWFAAGHR-PVGAELADRSEVIHGLVAGFLRSQ
MMAR_4185|M.marinum_M ITATLRMVRQR--PALAAWFASTRP-PIGGELAGQSEVIAGLAAAFLQSL
MUL_4487|M.ulcerans_Agy99 ITATLRMVRQR--PALAAWFASTRP-PIGGELAGQSEVIAGLAAAFLQSL
MAV_1403|M.avium_104 ITTTLRMVRGN--PALASWFAVTRP-PIAGELAEHSEVITALAAAFVGSL
Rv1255c|M.tuberculosis_H37Rv ITTTLRMVRDN--PALAAWFTTTRP-PIGGEMAGRSEVIAALAAAFLNSL
MLBr_02568|M.leprae_Br4923 IVSFLRYIDANRASWIVMYIQAMSLPAFAQTVREARDQITELVAGLMRVG
: * : . : . : . :
Mb1805c|M.bovis_AF2122/97 G----------------GDPQGAKWVVRVVLSLMYWPAENDEAERRLVEK
MAB_1938c|M.abscessus_ATCC_199 ------------------DETAAQWISRVVMALLFWPTSDPAQESEMIER
Mflv_2228|M.gilvum_PYR-GCK G-PDDADARASGATGEGSVAHRSRWLVRVMTSLMLFPGHDEADERAMLEE
Mvan_4466|M.vanbaalenii_PYR-1 G-PDDAEL----------VAHRARWLVRVMTSLMLFPGHDETDERAMLEE
MSMEG_5040|M.smegmatis_MC2_155 GSPTDADD-------DAAVGRRARWLVRVMVSLLVFPGVDEADERTMLAD
TH_2228|M.thermoresistible__bu GFDDDG--------------RRARWLVRVIVSLLLFPGRDEAEERALLED
MMAR_4185|M.marinum_M G-PEAPAA----------VERRARWTVRVIVSLLMFPGQDEADERAMLEE
MUL_4487|M.ulcerans_Agy99 G-PEAPAA----------VERRARWTVRVIVSLLMFPGQDEADERAMLEE
MAV_1403|M.avium_104 G-PEEPAV----------VERRARWVVRVITSLLLFPGHDEADERAMIEE
Rv1255c|M.tuberculosis_H37Rv G-PDDPTT----------VERRARWVVRMLTSLLMFPGRDEADERAMIAE
MLBr_02568|M.leprae_Br4923 TRSSRPDH------EYEIMAGALVGAGEAMATRLTTGDIAVDEAAELMIN
. : : : ::
Mb1805c|M.bovis_AF2122/97 YVAPAFAEQS------------------------------
MAB_1938c|M.abscessus_ATCC_199 FVYPVLKDATASRVSASATSESTISPARTATFDSSEDDSP
Mflv_2228|M.gilvum_PYR-GCK FVVPIVLPRRQADQATQ-----------------------
Mvan_4466|M.vanbaalenii_PYR-1 FVVPTVLPSRHAAR--------------------------
MSMEG_5040|M.smegmatis_MC2_155 FVAPLVVG-AQAAQ--------------------------
TH_2228|M.thermoresistible__bu FVVPAVAQATE-----------------------------
MMAR_4185|M.marinum_M FVAPVVTPFPTRR---------------------------
MUL_4487|M.ulcerans_Agy99 FVAPVVTPFPTRR---------------------------
MAV_1403|M.avium_104 FVVPIVTPVSARG---------------------------
Rv1255c|M.tuberculosis_H37Rv FVVPIVTPASAAARKAGHPGPE------------------
MLBr_02568|M.leprae_Br4923 LFWHGLKGAPADRDFGPSIVAG------------------
. .