For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRAVVIDGAGSVRVNTQPDPALPGPDGVVVAVTAAGICGSDLHFYEGEYPFTEPVALGHEAVGTIVEAGP QVRTVGVGDLVMVSSVAGCGVCPGCEPMIQSCASPAR
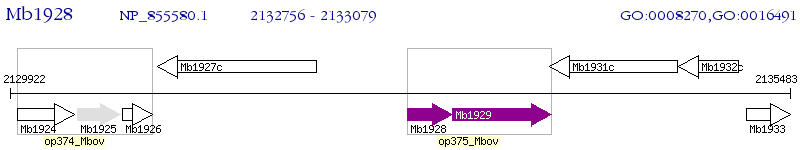
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1928 | - | - | 100% (107) | dehydrogenase |
| M. bovis AF2122 / 97 | Mb3113 | adhD | 9e-14 | 46.58% (73) | zinc-type alcohol dehydrogenase AdhD |
| M. bovis AF2122 / 97 | Mb2283 | adhE2 | 7e-13 | 45.95% (74) | zinc-dependent alcohol dehydrogenase AdhE2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4517 | - | 4e-32 | 62.11% (95) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv1895 | - | 3e-53 | 100.00% (96) | dehydrogenase |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 1e-13 | 45.95% (74) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_0983c | - | 3e-14 | 45.12% (82) | zinc-containing alcohol dehydrogenase |
| M. marinum M | MMAR_2790 | - | 5e-42 | 80.00% (95) | dehydrogenase |
| M. avium 104 | MAV_2804 | - | 8e-40 | 72.63% (95) | zinc-binding dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0671 | - | 1e-17 | 43.75% (96) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. thermoresistible (build 8) | TH_2319 | - | 4e-17 | 43.16% (95) | S-(hydroxymethyl)glutathione dehydrogenase |
| M. ulcerans Agy99 | MUL_2962 | - | 1e-41 | 78.95% (95) | dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1841 | - | 4e-33 | 65.26% (95) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01784|M.leprae_Br4923 ------MSQTVRGVISRKKDEPVELVDIVIPDPGPGEAVVDVTACGVCHT
MAB_0983c|M.abscessus_ATCC_199 MRIRSAVLHAAPIERPYRDTHPLKVIELELDPPGPGEVLVRIEAAGLCHS
Mflv_4517|M.gilvum_PYR-GCK ----------MRTVVVDSQRSIRVDTRPDPRLPGPDGVIVDVTASAICGS
Mvan_1841|M.vanbaalenii_PYR-1 ----------MRAVVIDSERRIRVETRPDPQLPGPDGAIVEVTATAICGS
Mb1928|M.bovis_AF2122/97 ----------MRAVVIDGAGSVRVNTQPDPALPGPDGVVVAVTAAGICGS
Rv1895|M.tuberculosis_H37Rv ----------MRAVVIDGAGSVRVNTQPDPALPGPDGVVVAVTAAGICGS
MMAR_2790|M.marinum_M ----------MRTVVIDGPGSIRVDNRPDPALPGSDGVIVAVSAAGICGS
MUL_2962|M.ulcerans_Agy99 ----------MRTVVIDGPGSIRVDNRPDPALPGSDGVIVVVSAAGICGS
MAV_2804|M.avium_104 ----------MRTVVVDGPRSIRVDTRPDPVLPGPDGAIVEVSAAGICGS
MSMEG_0671|M.smegmatis_MC2_155 ----------MRAMVYRGPYKIRLEDKPIPAIEHPNDAIVRVTKAAICGS
TH_2319|M.thermoresistible__bu -------------VTWHGRRKVSVDTVPDPAIMEPTDAIIRVTSTNICGS
. .:: : :* :
MLBr_01784|M.leprae_Br4923 DLTYREGGINDRYP-FLLGHEAAGTVEVVGPGVTAVEPGDFVILNWRAVC
MAB_0983c|M.abscessus_ATCC_199 DLSVVDGNRIRPVP-MALGHEAAGVIAAVGPGVRDVSPGDHVVLVYVPSC
Mflv_4517|M.gilvum_PYR-GCK DLHFLDGHYPIDQP-VPIGHEAVGVVVETGSDVTRFTNGDRVLVSSVTGC
Mvan_1841|M.vanbaalenii_PYR-1 DLHFLEGHYPIDQP-VSIGHEAVGVVVETGTTVAGFKVGDRVLVSSVAGC
Mb1928|M.bovis_AF2122/97 DLHFYEGEYPFTEP-VALGHEAVGTIVEAGPQVRTVGVGDLVMVSSVAGC
Rv1895|M.tuberculosis_H37Rv DLHFYEGEYPFTEP-VALGHEAVGTIVEAGPQVRTVGVGDLVMVSSVAGC
MMAR_2790|M.marinum_M DLHFYEGEYPLAEP-VALGHEAVGTVVEKGPDVHTVEVGDQVMVSSVAGC
MUL_2962|M.ulcerans_Agy99 DLHFYEGEYPLAEP-VALGHEAVGTVVEKGPDVHTVEVGDQVMVSSVAGC
MAV_2804|M.avium_104 DLHFYEADFPMPDP-IALGHEAIGTVVEAGPQVRNVKVGDQVMVSSVAGC
MSMEG_0671|M.smegmatis_MC2_155 DLHLYHGMMPDTRVGTTFGHEFIGVVEKVGPSVQNLSVGDRVMVPFNIFC
TH_2319|M.thermoresistible__bu DLHLYEVLSAFMSPGDILGHEAMGVVEEVGPEVGALQAGDRVVIPFNISC
** . :*** *.: *. * . ** *:: *
MLBr_01784|M.leprae_Br4923 GQCRACKRGRPSYCFDTFNAQQKMTLIDG---------TELTPALGIGAF
MAB_0983c|M.abscessus_ATCC_199 GACRYCSSGRPALCPDAAAANGRGDLIRGGTRLHDLDGTEVRHHLGVSGF
Mflv_4517|M.gilvum_PYR-GCK GRCAGCLTLDPVKCVHGPQIFGTG-------------------LLGGAQS
Mvan_1841|M.vanbaalenii_PYR-1 GRCAGCRTQDPVKCVQGPQIFGSG-------------------LLGGAQA
Mb1928|M.bovis_AF2122/97 GVCPGCE----------PMIQSC---------------------------
Rv1895|M.tuberculosis_H37Rv GVCPGCETHDPVMCFSGPMIFGAG-------------------VLGGAQA
MMAR_2790|M.marinum_M GACAGCATRDPAMCVSGPQIFGSG-------------------ALGGAQA
MUL_2962|M.ulcerans_Agy99 GACAGCATRDPAMCVSGPQIFGSG-------------------ALGGAQA
MAV_2804|M.avium_104 GSCAGCATRDPVLCHSGFQIFGGG-------------------VLGGAQA
MSMEG_0671|M.smegmatis_MC2_155 GSCYFCARGLYSNCHNVNPNATAVGGIYG--------YSHTCGGYDGGQA
TH_2319|M.thermoresistible__bu GTCWMCAQGLQSQCETTQNRDQGTGAALFG-------YSKLYGEVAGGQA
* * *
MLBr_01784|M.leprae_Br4923 ADKTLVHSGQCTKVDPAADPAVAGLLGCGVMAGLGAAINTAAVSRDDTVA
MAB_0983c|M.abscessus_ATCC_199 ADHAVVDRNSVVVIDENVPLDTAALFGCAMLTGFGAVTHTASVRPGDSVA
Mflv_4517|M.gilvum_PYR-GCK DLLAVPAADFQLLAVPDGIGTEEALLLTDNLPTGWAAAKRADIPVGGTVA
Mvan_1841|M.vanbaalenii_PYR-1 DLMAVPAADFQLLAIPDGIDTEQALLLTDNLATGWAAAKRADIPVGGTVA
Mb1928|M.bovis_AF2122/97 ---ASPAR------------------------------------------
Rv1895|M.tuberculosis_H37Rv DLLAVPAADFQVLKIPEGITTEQALLLTDNLATGWAAAQRADISFGSAVA
MMAR_2790|M.marinum_M DLLAVPAADFQVLKIPAAITTEQALLLTDNLATGWAAARRADILFGATVA
MUL_2962|M.ulcerans_Agy99 DLLAVPATDFQVLKIPAAITTEQALLLTDNLATGWAAARRADIPFGATVA
MAV_2804|M.avium_104 DLLAVPAADFQLLKIPEGISTEQALLLTDNLATGWAAAQRADIPFGGTVA
MSMEG_0671|M.smegmatis_MC2_155 EFVRVPFADVGPSIIPDRLDDEDALLLTDALATGYFGAQLGNIAEGDTVV
TH_2319|M.thermoresistible__bu EYLRVPQAQYTHITVPHDGPDDRYVYLSDVLPTAWQGVEYAAVPDGGTLV
MLBr_01784|M.leprae_Br4923 VIGCGGVGDAAISGAALVGANRIIAVDIDDTKLEWARTFG-ATHTVNALE
MAB_0983c|M.abscessus_ATCC_199 VLGLGGVGLAVVMSAAAAGAGEVLAIDPVPAKRELALELG-ATSACAPDE
Mflv_4517|M.gilvum_PYR-GCK VIGAGAVGQCALRSALALGAARVFAVDPVVARRERAAAAG-ATPMDSPAA
Mvan_1841|M.vanbaalenii_PYR-1 VIGAGAVGQCALRSAYALGAATVFAVDPVAARRDRAAAAG-ARAVPAPAA
Mb1928|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv VIGLGAVGLCALRSAFIHGAATVFAVDRVKGRLQRAATWG-ATPIPSPAA
MMAR_2790|M.marinum_M VIGLGAVGLCAVRSALAQGAATVFAVDRVQGRLQRASEWG-ATAVASPAA
MUL_2962|M.ulcerans_Agy99 VIGLGAVGLCAVRSALAQGAATVFAVDRVQGRLQRAAEWG-ATAVASPAA
MAV_2804|M.avium_104 VIGLGAVGLCALRSALFQGAATVFAVDQVDGRLARAARWG-ATPLKAPAL
MSMEG_0671|M.smegmatis_MC2_155 VFGAGPVGLFAAKSAWLMGAGRVIVIDHLEYRLEKARTFAHAETINFVDC
TH_2319|M.thermoresistible__bu VLGLGPIGSMACRIAAHRGNCRVIGVDRVPERIEKVRPYC--ADVLNVDT
MLBr_01784|M.leprae_Br4923 LEVVKTIQDLTSGFGVDVVIDTVGRPET---------------------W
MAB_0983c|M.abscessus_ATCC_199 APG-----------GHDWVFEVVGSAAV---------------------L
Mflv_4517|M.gilvum_PYR-GCK QTILEATD----GLGADSVIDAVGTDAS---------------------I
Mvan_1841|M.vanbaalenii_PYR-1 AAILEATG----GLGVDSVIDAVGTDTS---------------------L
Mb1928|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv ETILAATR----GRGADSVIDAVGTDAS---------------------M
MMAR_2790|M.marinum_M EAIVAATG----GRGADSVIDAVGTDAS---------------------M
MUL_2962|M.ulcerans_Agy99 EAIVAATS----GRGADSVIDAVDTDAS---------------------M
MAV_2804|M.avium_104 EAILAATG----GRGADAVIDAVATDAS---------------------L
MSMEG_0671|M.smegmatis_MC2_155 DDIVVKMKRTTDHLGADVAIDAAGAEADGNFLQHVTATKLKLQGGSPIAL
TH_2319|M.thermoresistible__bu DDVEAAVREQTGGXXVQTPPDPRRQSADCEN---HADQHGRPEHPPEELH
MLBr_01784|M.leprae_Br4923 KQAFYARDLAGTVVLVGVPTPDMRLDMPLLDFFSHGGSLKSS--------
MAB_0983c|M.abscessus_ATCC_199 EQAYALTGRGGGTVSVGLPHPDARISLPALNIVAEGRSILGS--------
Mflv_4517|M.gilvum_PYR-GCK DDALAGVRTGGTVSVVGVHDLTP-YPLPALVCLLRSLTLR----------
Mvan_1841|M.vanbaalenii_PYR-1 DDALACVRTGGTVSIVGVHDLQP-YPLPALVCLLRSLTIR----------
Mb1928|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv SDALNAVRPGGTVSVVGVHDLQP-FPVPALTCLLRSITLR----------
MMAR_2790|M.marinum_M TDALNAVRAGGTVSVVGVHDLQP-FPLPALTCLLRSITLR----------
MUL_2962|M.ulcerans_Agy99 TDALNAVRAGGTVSVVGVHDLQP-FPLPALTCLLRSITLR----------
MAV_2804|M.avium_104 TEALNAVRPGGTVSVVGVHDMNP-FPLNALGCLIRSITLR----------
MSMEG_0671|M.smegmatis_MC2_155 NWAIDSVRKGGTVSVVGAYGPMF-SAVKFGDAFNKGLTLR----------
TH_2319|M.thermoresistible__bu HQADDQRQRRHERDAATAGSPPLPSPWPDRTTTHRHTWRRLGGMELLRDV
MLBr_01784|M.leprae_Br4923 --WYGDCLPERDFPTLVDLYLQGRLPLGKFVSERIGLGDVE---------
MAB_0983c|M.abscessus_ATCC_199 --YMGSAQPQQDIPAMLALWRAGRLPVEKLKSDDLPLSDIN---------
Mflv_4517|M.gilvum_PYR-GCK ---MTTAPVQQTWAELVPLLQAGRLSVDGIFTDAMPLAEAE---------
Mvan_1841|M.vanbaalenii_PYR-1 ---LTTAPVQQTWPELIPLLQAGRLSVDGIFTGALPLDDAE---------
Mb1928|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv ---MTMAPVQRTWPELIPLLQSGRLDVDGIFTTTLPLDEAA---------
MMAR_2790|M.marinum_M ---LTIAPVQRTWSELIPLLESGRLDVDGIFTTSLPLDEAA---------
MUL_2962|M.ulcerans_Agy99 ---LTIAPVQRTWSELIPLLESGRLDVDGIFTTSLPLDEAA---------
MAV_2804|M.avium_104 ---MTTAPVQRTWPELIPLLQSGRLDVDGIFTTTMPLDEAA---------
MSMEG_0671|M.smegmatis_MC2_155 ---MNQCPVKREWPRLLSHIKNGYLKPNDIVTHRIPLEHIA---------
TH_2319|M.thermoresistible__bu VVLLHIVGFAVTFGAWTAEAVARRFRTTRLMDYGLLLSLITGLALAAPWP
MLBr_01784|M.leprae_Br4923 EAFHKIHGGKVLRSVVML--------------------------------
MAB_0983c|M.abscessus_ATCC_199 IALDALAEGQAVRQILRPGVSA----------------------------
Mflv_4517|M.gilvum_PYR-GCK AAYAAAFSRSGEHLKVHLVP------------------------------
Mvan_1841|M.vanbaalenii_PYR-1 RAYAAAFSRSAEHLKVQLIP------------------------------
Mb1928|M.bovis_AF2122/97 --------------------------------------------------
Rv1895|M.tuberculosis_H37Rv KGYATARARSGEELRFCLRPDSRDVLGAHETVDLYVHVRRCQSVADLQLE
MMAR_2790|M.marinum_M KGYSIAGSRSGNDVKVLLKV------------------------------
MUL_2962|M.ulcerans_Agy99 KGYSIAGSRSGNDVKVLLKV------------------------------
MAV_2804|M.avium_104 IGYATAGSRSGDDVKILLKP------------------------------
MSMEG_0671|M.smegmatis_MC2_155 EGYHIFSAKLDDCIKPIIVPTAS---------------------------
TH_2319|M.thermoresistible__bu AGIVLNYPKIGLKLVLLVALGAVLGIGGAHQRRTGNPVPRPLFVTAGVLS
MLBr_01784|M.leprae_Br4923 -------------
MAB_0983c|M.abscessus_ATCC_199 -------------
Mflv_4517|M.gilvum_PYR-GCK -------------
Mvan_1841|M.vanbaalenii_PYR-1 -------------
Mb1928|M.bovis_AF2122/97 -------------
Rv1895|M.tuberculosis_H37Rv GAADGVDGPSMLN
MMAR_2790|M.marinum_M -------------
MUL_2962|M.ulcerans_Agy99 -------------
MAV_2804|M.avium_104 -------------
MSMEG_0671|M.smegmatis_MC2_155 -------------
TH_2319|M.thermoresistible__bu FAAAAVAVLW---