For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIRSAVLHAAPIERPYRDTHPLKVIELELDPPGPGEVLVRIEAAGLCHSDLSVVDGNRIRPVPMALGHEA AGVIAAVGPGVRDVSPGDHVVLVYVPSCGACRYCSSGRPALCPDAAAANGRGDLIRGGTRLHDLDGTEVR HHLGVSGFADHAVVDRNSVVVIDENVPLDTAALFGCAMLTGFGAVTHTASVRPGDSVAVLGLGGVGLAVV MSAAAAGAGEVLAIDPVPAKRELALELGATSACAPDEAPGGHDWVFEVVGSAAVLEQAYALTGRGGGTVS VGLPHPDARISLPALNIVAEGRSILGSYMGSAQPQQDIPAMLALWRAGRLPVEKLKSDDLPLSDINIALD ALAEGQAVRQILRPGVSA*
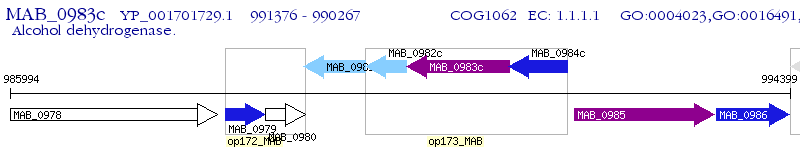
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0983c | - | - | 100% (369) | zinc-containing alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2084 | - | 5e-62 | 38.72% (359) | zinc-type alcohol dehydrogenase AdhD |
| M. abscessus ATCC 19977 | MAB_1869c | - | 5e-57 | 32.58% (353) | zinc-dependent alcohol dehydrogenase AdhE2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0167c | adhE1 | 1e-100 | 53.02% (364) | putative zinc-type alcohol dehydrogenase (e subunit) adhE |
| M. gilvum PYR-GCK | Mflv_1486 | - | 3e-61 | 36.67% (360) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv0162c | adhE1 | 1e-100 | 53.02% (364) | zinc-type alcohol dehydrogenase E subunit |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 3e-60 | 35.98% (353) | putative alcohol dehydrogenase (Zn dependent) |
| M. marinum M | MMAR_0405 | adhE | 1e-101 | 51.91% (366) | zinc-type alcohol dehydrogenase (E subunit) AdhE |
| M. avium 104 | MAV_5022 | - | 1e-100 | 53.19% (361) | oxidoreductase, zinc-binding dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_0127 | - | 1e-100 | 51.92% (364) | oxidoreductase, zinc-binding dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_3466 | - | 1e-66 | 48.91% (276) | PUTATIVE Alcohol dehydrogenase GroES domain protein |
| M. ulcerans Agy99 | MUL_1292 | adhE2 | 9e-60 | 35.13% (353) | zinc-dependent alcohol dehydrogenase AdhE2 |
| M. vanbaalenii PYR-1 | Mvan_5270 | - | 1e-62 | 37.78% (360) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0167c|M.bovis_AF2122/97 -MPAVQPWLYSNMPAIRGAVLDQIGVPRPYWRSKPISVVELHLDPPDRGE
Rv0162c|M.tuberculosis_H37Rv -MPAVQPWLYSNMPAIRGAVLDQIGVPRPYWRSKPISVVELHLDPPDRGE
MMAR_0405|M.marinum_M MELGLQPWLYSNMIHIRGAVLERIGSPRPYAQSRPISVVDLDLDPPGDDE
MAV_5022|M.avium_104 -------------------MLQRIGAPRPYARSRPISITDVELAPPGRDE
MSMEG_0127|M.smegmatis_MC2_155 -------------MKIRGAVLERIGAPRPYAESRPLTISELDLADPGPDE
TH_3466|M.thermoresistible__bu --------------------------------------------------
MAB_0983c|M.abscessus_ATCC_199 -------------MRIRSAVLHAAPIERPYRDTHPLKVIELELDPPGPGE
MLBr_01784|M.leprae_Br4923 -------------------MSQTVRGVISRKKDEPVELVDIVIPDPGPGE
MUL_1292|M.ulcerans_Agy99 -------------------MSQTVRGVISRKKGEPVELVDIVVPDPGPGE
Mflv_1486|M.gilvum_PYR-GCK ---------------------MKTNAAILWEYGGDWTVEEIDLDPPGDGE
Mvan_5270|M.vanbaalenii_PYR-1 ---------------------MKTNAAILWEYGGDWTVEEIDLDPPGDGE
Mb0167c|M.bovis_AF2122/97 VLVRIEAAGVCHSDLSVVDGTRVRPVPILLGHEAAGIVEQVGDGVDGVAV
Rv0162c|M.tuberculosis_H37Rv VLVRIEAAGVCHSDLSVVDGTRVRPVPILLGHEAAGIVEQVGDGVDGVAV
MMAR_0405|M.marinum_M VLVRIEAAGVCHSDLSVVDGNRVRPVPMLLGHEAAGIVERVGDRVDDLAV
MAV_5022|M.avium_104 VLVRIEAAGICHSDLSVVDGNRVRPVPMLLGHEAAGIVERVGPGVADLAV
MSMEG_0127|M.smegmatis_MC2_155 LLVRIESAGLCHSDLSVVDGNRVRPVPMLLGHEAAGVVETPG---ADLET
TH_3466|M.thermoresistible__bu --------------------------------------------------
MAB_0983c|M.abscessus_ATCC_199 VLVRIEAAGLCHSDLSVVDGNRIRPVPMALGHEAAGVIAAVGPGVRDVSP
MLBr_01784|M.leprae_Br4923 AVVDVTACGVCHTDLTYREGGINDRYPFLLGHEAAGTVEVVGPGVTAVEP
MUL_1292|M.ulcerans_Agy99 AVVDISACGVCHTDLTYREGGINDEYPFLLGHEAAGTVEAVGPGVTNVEP
Mflv_1486|M.gilvum_PYR-GCK VLVSWQATGLCHSDEHIRTGDLPAPLPLIGGHEGAGIVREVGAGVVGLAP
Mvan_5270|M.vanbaalenii_PYR-1 VLVSWEATGLCHSDEHIRTGDLPAPLPLIGGHEGAGIVREVGPGVVGLAP
Mb0167c|M.bovis_AF2122/97 GQRVVLVFLPRCGQCAACATDGRTPCEPGSAANKAGTLLGGGIRLS-RGG
Rv0162c|M.tuberculosis_H37Rv GQRVVLVFLPRCGQCAACATDGRTPCEPGSAANKAGTLLGGGIRLS-RGG
MMAR_0405|M.marinum_M GQRVVLVFLPRCGHCPSCATEGITPCAPGSATNAAGTLLGGGIRIS-RSG
MAV_5022|M.avium_104 GARVVLVFLPRCGRCAACATEGLTPCEPGSAANGAGTLLGGDIRLS-RAG
MSMEG_0127|M.smegmatis_MC2_155 GQRVVMTFLPRCGSCPACGTDGLTPCGPGSVANGAGTLMNGDIRLR-RDG
TH_3466|M.thermoresistible__bu ---VVMTFLPRCGHCPACATDGLTPCEPGSAANTAGTLLSGAIRLT-RDG
MAB_0983c|M.abscessus_ATCC_199 GDHVVLVYVPSCGACRYCSSGRPALCPDAAAANGRGDLIRGGTRLHDLDG
MLBr_01784|M.leprae_Br4923 GDFVILNWRAVCGQCRACKRGRPSYCFDTFNAQQKMTLIDG---------
MUL_1292|M.ulcerans_Agy99 GDFVILNWRAVCGQCRACKRGRPSYCFDTFNAEQKMTLTDG---------
Mflv_1486|M.gilvum_PYR-GCK GDHVVASFLPACGRCRFCSTGHQNLCDLG-AMIMAGTQLDGTYRRH-ING
Mvan_5270|M.vanbaalenii_PYR-1 GDHVVASFLPACGRCRFCSTGHQNLCDLG-AMIMAGTQLDGTYRRH-VRG
*: : . ** * * * *
Mb0167c|M.bovis_AF2122/97 RPVYHHLGVSGFATHVVVNRASVVPVPHEVPPTVAALLGCAVLTGGGAVL
Rv0162c|M.tuberculosis_H37Rv RPVYHHLGVSGFATHVVVNRASVVPVPHEVPPTVAALLGCAVLTGGGAVL
MMAR_0405|M.marinum_M QPIYHHLGVSGFATHAVVNRASAVAVPDEVPPQVAALLGCAVLTGGGAVL
MAV_5022|M.avium_104 RPVYHHLGVSGFATHAVVNRASAVEVPPDVPAEVAALLGCAVLTGGGAVL
MSMEG_0127|M.smegmatis_MC2_155 EPVFHHLGVSGFATHAVVDRRSIVPVPDDVPATVAALLGCAVLTGGGAVL
TH_3466|M.thermoresistible__bu QPVHHHLGISGFASHAVVDRRSVVPVDSDVPPSVAALLGCAVLTGGGAVL
MAB_0983c|M.abscessus_ATCC_199 TEVRHHLGVSGFADHAVVDRNSVVVIDENVPLDTAALFGCAMLTGFGAVT
MLBr_01784|M.leprae_Br4923 TELTPALGIGAFADKTLVHSGQCTKVDPAADPAVAGLLGCGVMAGLGAAI
MUL_1292|M.ulcerans_Agy99 TELTPALGIGAFADKTLVHSGQCTKVDPAADPAVAGLLGCGVMAGLGAAV
Mflv_1486|M.gilvum_PYR-GCK QDVGVMALVGTFSQYGTVPEASIVKIDDDIPLSRACLLGCGVTTGWGSAV
Mvan_5270|M.vanbaalenii_PYR-1 QDVGVMALVGTFSQYGTVPEASIVKIDDDIPLSRACLLGCGVTTGWGSAV
: :. *: * . . : * *:**.: :* *:.
Mb0167c|M.bovis_AF2122/97 NVGDPQPGQSVAVVGLGGVGMAAVLTALTYTDVRVVAVDQLPEKLSAAKA
Rv0162c|M.tuberculosis_H37Rv NVGDPQPGQSVAVVGLGGVGMAAVLTALTYTDVRVVAVDQLPEKLSAAKA
MMAR_0405|M.marinum_M NVGKPVAGQSVAVVGLGGVGMAAVLTALTQDGVAVFGVDQLPQKLSAAEA
MAV_5022|M.avium_104 NVGRPAPGQTVAVVGLGGVGMAALLTALTYDGVPVVAVDQLPEKLAAARR
MSMEG_0127|M.smegmatis_MC2_155 NVGRPKPGQTVVVVGLGGVGMAAALTALAHDDVRVVGVDQLSDKLARAAE
TH_3466|M.thermoresistible__bu NVGRPRPGQRVIVVGLGGVGLAAVLTALAHPDVRVVGVDQRTDKLHTART
MAB_0983c|M.abscessus_ATCC_199 HTASVRPGDSVAVLGLGGVGLAVVMSAAAAGAGEVLAIDPVPAKRELALE
MLBr_01784|M.leprae_Br4923 NTAAVSRDDTVAVIGCGGVGDAAISGAALVGANRIIAVDIDDTKLEWART
MUL_1292|M.ulcerans_Agy99 NTGGVTRDDTVAVIGCGGVGDAAIAGAALVGAKKIIAVDTDNTKLEWARA
Mflv_1486|M.gilvum_PYR-GCK NTADVSPGDTVVIIGLGGIGSGAIQGARLAGAEKIVVVEPVETKRDLAFR
Mvan_5270|M.vanbaalenii_PYR-1 NTADVSPGDTVVIIGLGGIGSGAIQGARLAGAEKIVVIEPVESKRELAFR
:.. .: * ::* **:* .. * :. :: * *
Mb0167c|M.bovis_AF2122/97 LGAHEIYTP--------QQATAGGVKAAVVVEAVGHPA--ALHTAIGLTA
Rv0162c|M.tuberculosis_H37Rv LGAHEIYTP--------QQATAGGVKAAVVVEAVGHPA--ALHTAIGLTA
MMAR_0405|M.marinum_M LGASATYTP--------QQAAAAGVKADVVIEAAGHPA--ALETAIGLTA
MAV_5022|M.avium_104 LGAHQAYTP--------QQAVEARVKAAVVIEAVGHPA--ALQTAIDLTA
MSMEG_0127|M.smegmatis_MC2_155 LGVHETFTP--------EQAAD--LRAEVVIEAAGHPA--ALETAIGLTG
TH_3466|M.thermoresistible__bu LGAHEAYTP--------DQAVDANLKAQVVIEAAGHPD--ALATAIQLTA
MAB_0983c|M.abscessus_ATCC_199 LGATSACAP--------DEAPGG---HDWVFEVVGSAA--VLEQAYALTG
MLBr_01784|M.leprae_Br4923 FGATHTVNALELEVVKTIQDLTSGFGVDVVIDTVGRPE--TWKQAFYARD
MUL_1292|M.ulcerans_Agy99 FGATHTVNARELDVVETIQDLTGGFGVDVVIDAVGRPE--TWKQAFYARD
Mflv_1486|M.gilvum_PYR-GCK FGATHFVTS-MEEATALVAELTRGVMADSAILTVGLLAGSMLEEAANIIR
Mvan_5270|M.vanbaalenii_PYR-1 FGATHFVTS-MEEATALVADLTRGVMADSAILTVGLLEGSMLEEAANIIR
:*. . .. ..* *
Mb0167c|M.bovis_AF2122/97 PGGRTITVGLP-PPDVRISLSPLDFVTEGRSLIGSYLGSAVPSHDIPRFV
Rv0162c|M.tuberculosis_H37Rv PGGRTITVGLP-PPDVRISLSPLDFVTEGRSLIGSYLGSAVPSHDIPRFV
MMAR_0405|M.marinum_M PGGRTITVGLP-PPDARISVAPLGFVAEGRSLIGSYLGSAVPSRDIPRFV
MAV_5022|M.avium_104 PGGRTITVGLP-PPDARITVSPLGFVAEGRSLIGSYLGSAVPSRDIPRFV
MSMEG_0127|M.smegmatis_MC2_155 PGGRTITVGLP-RPDARITVSPLGFVAEGRSLIGSYLGSAVPSRDIPRFV
TH_3466|M.thermoresistible__bu PGGRTITVGLP-PPDARLTVSPLGLVAEGRSLIGSYLGSAVPARDIPRFV
MAB_0983c|M.abscessus_ATCC_199 RGGGTVSVGLP-HPDARISLPALNIVAEGRSILGSYMGSAQPQQDIPAML
MLBr_01784|M.leprae_Br4923 LAGTVVLVGVP-TPDMRLDMPLLDFFSHGGSLKSSWYGDCLPERDFPTLV
MUL_1292|M.ulcerans_Agy99 LAGTVVLVGVP-TPDMRLDMPLVDFFSRGGALKSSWYGDCLPERDFPTLI
Mflv_1486|M.gilvum_PYR-GCK KGGAVVMTALSTMADNQPTLGMMMFTLFQKRLLGSLYGEANPRADIPRLL
Mvan_5270|M.vanbaalenii_PYR-1 KGGAVVMTALSTMADNQPTLGMMMFTLFQKRLLGSLYGEANPRADIPRLL
.* .: ..:. .* : : : : : .* *.. * *:* ::
Mb0167c|M.bovis_AF2122/97 SLWQSGRLPVESLVTSTIRLDDINEAMDHLADGIAVRQLISFTGDL
Rv0162c|M.tuberculosis_H37Rv SLWQSGRLPVESLVTSTIRLDDINEAMDHLADGIAVRQLISFTGDL
MMAR_0405|M.marinum_M SLWQSGRLPVESLVSATIRLDDLNEAMDHLADGTAVRQIIVF----
MAV_5022|M.avium_104 ELWRAGRLPVESLVSSTIRLEDINEAMDHLADGTAVRQLIRFDA--
MSMEG_0127|M.smegmatis_MC2_155 ELWRAGRLPVESLVSSTIALDDINAGMDELAEGRAVRQVIDFA---
TH_3466|M.thermoresistible__bu DLWRSGRLPVEQLVSSTITLDELNEGLDRLADGAAVRQLITLPG--
MAB_0983c|M.abscessus_ATCC_199 ALWRAGRLPVEKLKSDDLPLSDINIALDALAEGQAVRQILRPGVSA
MLBr_01784|M.leprae_Br4923 DLYLQGRLPLGKFVSERIGLGDVEEAFHKIHGGKVLRSVVML----
MUL_1292|M.ulcerans_Agy99 DLYLQGRLPLDKFVSEQIGLGGIEEAFEKMHAGKVLRSVVVL----
Mflv_1486|M.gilvum_PYR-GCK SLYREGKLLLDETVTHEYKLAEVNEGYEDMREGRNIRGVILHDH--
Mvan_5270|M.vanbaalenii_PYR-1 SLYREGKLLLDETVTHEYKLAEVNEGYEDMRAGRNIRGVILHDH--
*: *:* : . : * :: . . : * :* ::