For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SPPPAATALAERKRTATRQRIATAAAQLVADLGLVAATVERIADTADISRATFFRYFNSKEDAVAEGMTA HWLDRIATALAAQPEHQSAVEAVVGAFGGLAVGFAEMEDQVRELATLTRSSETLEAWTLHIYVRYESAIA DLIAPRIPNLVPQDPRPRLVGALAMAVVRIALDDWLGQGGSLPDRVQQGLAAIVIA*
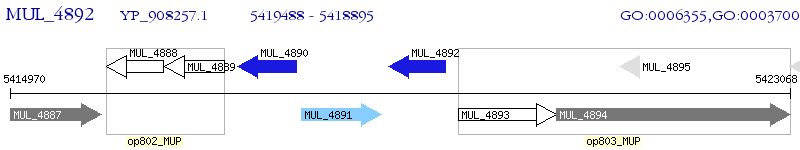
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4892 | - | - | 100% (197) | TetR family transcriptional regulator |
| M. ulcerans Agy99 | MUL_3793 | - | 5e-09 | 26.82% (179) | putative regulatory protein |
| M. ulcerans Agy99 | MUL_2714 | - | 9e-09 | 32.47% (194) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3860c | - | 2e-08 | 34.75% (118) | transcriptional regulatory protein TetR-family |
| M. gilvum PYR-GCK | Mflv_4072 | - | 2e-60 | 58.88% (197) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3830c | - | 2e-08 | 34.75% (118) | transcriptional regulatory protein TetR-family |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0248c | - | 4e-10 | 29.76% (168) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0222 | - | 1e-105 | 98.48% (197) | TetR family transcriptional regulator |
| M. avium 104 | MAV_2657 | - | 8e-15 | 31.07% (177) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1420 | - | 9e-11 | 28.40% (169) | transcriptional regulatory protein |
| M. thermoresistible (build 8) | TH_3524 | - | 3e-08 | 27.88% (165) | PUTATIVE - |
| M. vanbaalenii PYR-1 | Mvan_2270 | - | 5e-64 | 65.96% (188) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4892|M.ulcerans_Agy99 ---------MSPPPAATALAERKRTATRQRIATAAAQLVADLG-LVAATV
MMAR_0222|M.marinum_M ---------MSPPPAATSLAERKRTATRQRIATAAAQLVADLG-LAAATV
Mflv_4072|M.gilvum_PYR-GCK MRHSLIMDAVSGPKSGESLTVRRQRATKARIAAAAAEAVAAHG-LAGATI
Mvan_2270|M.vanbaalenii_PYR-1 ------MDVVSGVDSGESLTVRKQRATKARIAAAAAQSVIDHG-LAAATV
Mb3860c|M.bovis_AF2122/97 --------MVRPPQTARSER------TREALRQAALVRFLAQG-VEATSA
Rv3830c|M.tuberculosis_H37Rv --------MVRPPQTARSER------TREALRQAALVRFLAQG-VEATSA
MAV_2657|M.avium_104 --------MARGDRSPRDAHADR---VRNALLNAALNLFSTNG-YDETTT
TH_3524|M.thermoresistible__bu ---------VRGHRN------------REALIAAAIDLFTVNG-YEQTTV
MSMEG_1420|M.smegmatis_MC2_155 --------MSEGSRAGRRRS-----TTQDHIAGVAIDLFAARG-FDAVSV
MAB_0248c|M.abscessus_ATCC_199 ------MRPASGLREQKKSE------TKLALSRAALELALDRGDLAAVTV
: : .* * .:
MUL_4892|M.ulcerans_Agy99 ERIADTADISRATFFRYFNSKEDAVAEGMTAHWLDRIATALAAQPEHQSA
MMAR_0222|M.marinum_M ERIADTADISRATFFRYFNSKEDAVAEGMTAHWLDRIATALAAQPEHLSA
Mflv_4072|M.gilvum_PYR-GCK EHIAAAAEVGRATFFRYFSAKEDAVVEGMAAHWLDAIADAIAAQPPELAA
Mvan_2270|M.vanbaalenii_PYR-1 ERIAAAAEVGRATFFRYFSAKEDAVAEGMTAHWLDLITAAIAAQPPHLPA
Mb3860c|M.bovis_AF2122/97 EQIAEDAGVSLRTFYRHFRSKHDLLFADYDAG-LHWFRAALDARPADESI
Rv3830c|M.tuberculosis_H37Rv EQIAEDAGVSLRTFYRHFRSKHDLLFADYDAG-LHWFRAALDARPADESI
MAV_2657|M.avium_104 DQIAESVGVSPRTFFRYFPTKESVLFFG-EYDFIDAVSGVYLAQPEEASD
TH_3524|M.thermoresistible__bu EQIAEAAGVAPRTFFRHFAAKEDILFHGYADRLAEATRRFRAARSG--SL
MSMEG_1420|M.smegmatis_MC2_155 DDVAAAAGISRRTLFRYYASKSAIPWGDFDSH-LQHLQNLLRTLSSEVSL
MAB_0248c|M.abscessus_ATCC_199 DDIADAVRVSARTFRNYFTSKEDAVLFS-LRGIEDNAVALLRQRPQHEHV
: :* . :. *: .:: :* . . .
MUL_4892|M.ulcerans_Agy99 VEAVVGAFGGLAVGFAEMEDQVRELATLTRSSETLEAWTLHIYVRYESAI
MMAR_0222|M.marinum_M VEAVVGAFGGLAVGFAEMEDQVRELATLTRSSETLEAWTLHIYVRYESAI
Mflv_4072|M.gilvum_PYR-GCK DAAVVAAFQDLGHGFEAVSDQVRELAVMTRSSPVFSAWTLQLYLRYEAAI
Mvan_2270|M.vanbaalenii_PYR-1 GAALAAAFEDLGDGFDAISGQVRELATLTRSSPVFSAWTLQIYLRYESAI
Mb3860c|M.bovis_AF2122/97 IDSVQAAIFSFPYDVDAVT-KIASLRRGELEPSRIVRHMREVEADFADAI
Rv3830c|M.tuberculosis_H37Rv IDSVQAAIFSFPYDVDAVT-KIASLRRGELEPSRIVRHMREVEADFADAI
MAV_2657|M.avium_104 FEALANSFALLAPGLKRIRKRIAQYHDAVASSLVLLGRERKNHEANAETV
TH_3524|M.thermoresistible__bu WKALAEASEAVATAIAENPGIFLVRARMYGELPALRATMLRINDDWIDQM
MSMEG_1420|M.smegmatis_MC2_155 GDALREALLTFNTYADHEMAEHRQRMRVILETEELQAYSMTMYAGWRTVI
MAB_0248c|M.abscessus_ATCC_199 VDSLEAVMLDLAT--SEAFSTTVAVTRLSAQNPGLAAHDAARSEDVGTVV
:: . . : :
MUL_4892|M.ulcerans_Agy99 ADLIAPR---IPNLVPQDPRPRLVGALAMAVVRIALDDWLGQG----GSL
MMAR_0222|M.marinum_M ADLIAPR---IPNLVPQDPRPRLVGALAMAVVRIALDDWLGQG----GSL
Mflv_4072|M.gilvum_PYR-GCK ADLVGIR---MADPAPDDPRPRLLGALAMASVRIALDDWLVHG----GSL
Mvan_2270|M.vanbaalenii_PYR-1 AELLGPR---CPDPAPDDPRPRLLGALAMASVRIALDDWLVHG----GSL
Mb3860c|M.bovis_AF2122/97 QAQLRRRNCDIAGAPDARLHIAVTARCVAAAVFGAMEAWMLGSDRSLGEL
Rv3830c|M.tuberculosis_H37Rv QAQLRRRNCDIAGAPDARLHIAVTARCVAAAVFGAMEAWMLGSDRSLGEL
MAV_2657|M.avium_104 AQVIAER----RRMPAPDSECRLLAAVGMLLVERALNQWLSTPG---RSL
TH_3524|M.thermoresistible__bu TTEVARW---LGSDAAVDVRPRLAATVINGANRTAIDLWVSADGK--GDL
MSMEG_1420|M.smegmatis_MC2_155 AEYVADR----LGMSTTDLMPQTVAWLMLGVALSAYENWLADES---VSL
MAB_0248c|M.abscessus_ATCC_199 LTEVSRR---AGLDPDVDLYPRLVCNAAYAVLATVIQLAVSGSKLS-KDP
: . : : .
MUL_4892|M.ulcerans_Agy99 PDRVQQGLAAIVIA---------------------
MMAR_0222|M.marinum_M PDRVQQGLAAIVIA---------------------
Mflv_4072|M.gilvum_PYR-GCK PERVRTALASLHVA---------------------
Mvan_2270|M.vanbaalenii_PYR-1 PDRVRAALASIEVV---------------------
Mb3860c|M.bovis_AF2122/97 ARVCHVALESLRVGISDTWTTLTVSS---------
Rv3830c|M.tuberculosis_H37Rv ARVCHVALESLRVGISDTWTTLTVSS---------
MAV_2657|M.avium_104 DDLIRQEFAALPAVLK-------------------
TH_3524|M.thermoresistible__bu VELMADAVRLVRPTLDRIERESRTVANPVRRRDVV
MSMEG_1420|M.smegmatis_MC2_155 RGAIAAAFEVARPGLDALQPAH-------------
MAB_0248c|M.abscessus_ATCC_199 GALVAQGFQRLRDGLA-------------------
.