For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEGSRAGRRRSTTQDHIAGVAIDLFAARGFDAVSVDDVAAAAGISRRTLFRYYASKSAIPWGDFDSHLQH LQNLLRTLSSEVSLGDALREALLTFNTYADHEMAEHRQRMRVILETEELQAYSMTMYAGWRTVIAEYVAD RLGMSTTDLMPQTVAWLMLGVALSAYENWLADESVSLRGAIAAAFEVARPGLDALQPAH*
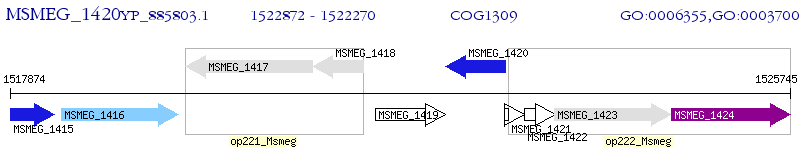
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1420 | - | - | 100% (200) | transcriptional regulatory protein |
| M. smegmatis MC2 155 | MSMEG_0363 | - | 8e-10 | 29.68% (155) | TetR-family protein regulatory protein |
| M. smegmatis MC2 155 | MSMEG_6862 | - | 5e-08 | 23.28% (189) | putative transcription regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0710c | - | 5e-72 | 68.88% (196) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_5062 | - | 1e-68 | 65.62% (192) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0691c | - | 5e-72 | 68.88% (196) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_02568 | - | 5e-08 | 24.53% (159) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3837 | - | 5e-59 | 57.65% (196) | transcriptional regulatory protein TetR |
| M. marinum M | MMAR_1019 | - | 5e-73 | 70.41% (196) | transcriptional regulator |
| M. avium 104 | MAV_4481 | - | 3e-72 | 71.73% (191) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_1899 | - | 4e-65 | 67.05% (176) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_0771 | - | 3e-73 | 70.41% (196) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1294 | - | 5e-69 | 66.32% (193) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0710c|M.bovis_AF2122/97 -MPHESRVGRRRSTTPHHISDVAIELFAAHGFTDVSVDDIARAAGIARRT
Rv0691c|M.tuberculosis_H37Rv -MPHESRVGRRRSTTPHHISDVAIELFAAHGFTDVSVDDIARAAGIARRT
MMAR_1019|M.marinum_M -MAHESRAGRRPSTTKRHIADVAIDLFAARTFAEVSVDDVAQAAGIARRT
MUL_0771|M.ulcerans_Agy99 -MAHESRAGRRPSTTKRHIADVAIDLFAARTFAEVSVDDVAQAAGIARRT
MAV_4481|M.avium_104 -MPQQ-RVGRRRSTTPHHITDVAIDLFTARGFAEVSVDDVAAAAGISRRT
MSMEG_1420|M.smegmatis_MC2_155 -MSEGSRAGRRRSTTQDHIAGVAIDLFAARGFDAVSVDDVAAAAGISRRT
Mflv_5062|M.gilvum_PYR-GCK MSAARPRVGRRRSTSWEHISDVAIDLFMARGFDDVSVDDVASAAGIARRT
Mvan_1294|M.vanbaalenii_PYR-1 MSAAKARVGRRRSTSWEHLSDVAIDLFMARGFDDVSVDDVAAAAGIARRT
TH_1899|M.thermoresistible__bu ---------------------VALDLFADRGFDEVSVDDVAAAAGIGRRT
MAB_3837|M.abscessus_ATCC_1997 MNAQPARVGRRPSTTRDEITAVAMTLFAQHGFDEVSVDDIAAAAGIARRT
MLBr_02568|M.leprae_Br4923 MAGGAKRIPR--AVREQQMLDAAVQMFSVNGYHKTSMDAIAAEAQISKPM
.*: :* . : .*:* :* * *.:
Mb0710c|M.bovis_AF2122/97 LFRYYASKNAIPWGDFSTHLAQLQGLL-DNIDSRIQLRDALRAALLAFNT
Rv0691c|M.tuberculosis_H37Rv LFRYYASKNAIPWGDFSTHLAQLQGLL-DNIDSRIQLRDALRAALLAFNT
MMAR_1019|M.marinum_M LFRYYASKNAIPWGDFDTHLAQLQDLL-ERIDGHVRLGKALREALLAFNT
MUL_0771|M.ulcerans_Agy99 LFRYYASKNAIPWGDFDTHLAQLQDLL-ERIDGHVRLGKALREALLAFNT
MAV_4481|M.avium_104 LFRYYASKNAIPWGDFDTHLARLRELL-DNVESRVPLGDALRTALLAFNT
MSMEG_1420|M.smegmatis_MC2_155 LFRYYASKSAIPWGDFDSHLQHLQNLL-RTLSSEVSLGDALREALLTFNT
Mflv_5062|M.gilvum_PYR-GCK LFRYYPSKSALPWGDFDAHLDHMRALL-ADLDPEVPIRDALRTALLAFND
Mvan_1294|M.vanbaalenii_PYR-1 LFRYYPSKSALPWGDFDAHLEHMRELL-DELDPSVPIRDALRTALLAFND
TH_1899|M.thermoresistible__bu LFRYYPSKNAIPWGDFDAHLDRMRDLL-DGIGPDVPIGAALRSMLLAFND
MAB_3837|M.abscessus_ATCC_1997 VFRYYSSKNAIVWGDFDSHLGVMRALL-DETADTVDTSTALRQAVLTFND
MLBr_02568|M.leprae_Br4923 LYLYYGSKEDLFGACLNREMSRFIAMVRADIDFTQSPKDLLRNTIVSFLR
:: ** **. : . :. .: : :: ** :::*
Mb0710c|M.bovis_AF2122/97 FDESETIRHRKRMRVILQTPELQAYSMTMYAGWREVIAKFVARRSGGKTT
Rv0691c|M.tuberculosis_H37Rv FDESETIRHRKRMRVILQTPELQAYSMTMYAGWREVIAKFVARRSGGKTT
MMAR_1019|M.marinum_M YDESETIRHRQRMRIILQTAELQAYSMTMYAGWRAVIAGFVARRLSVKPT
MUL_0771|M.ulcerans_Agy99 YDESETIRHRQRMRIILQTAELQAYSMTMYAGWRAVIAGFVARRLSVKPT
MAV_4481|M.avium_104 FDESETARHRQRMRVILQTAELQAYSMTMYAGWREVIAGFVARRCECKTT
MSMEG_1420|M.smegmatis_MC2_155 YADHEMAEHRQRMRVILETEELQAYSMTMYAGWRTVIAEYVADRLGMSTT
Mflv_5062|M.gilvum_PYR-GCK F--DDPDRHRQRMRLILETEALQAYSMTMYAGWRAVVAAFVADRLGVGGD
Mvan_1294|M.vanbaalenii_PYR-1 F--DDADRHRQRMRLILETEALQAYSMTMYAGWRAVVAAFVAGRLGVDPA
TH_1899|M.thermoresistible__bu FGEDETGRHRRRMRVILQTDALQAHSMTMYAGWRAVVADFVARRLGLAPS
MAB_3837|M.abscessus_ATCC_1997 FPVDETPRHRMRMRMILEVPALQAYSMLMYTGWRDVIAQFVARRSGTDPG
MLBr_02568|M.leprae_Br4923 YIDANRASWIVMYIQAMSLPAFAQTVREARDQITELVAGLMRVGTRSSRP
: : :. : ::* :
Mb0710c|M.bovis_AF2122/97 DFMPQTVAWTMLGVALSAYEHWLRDESVSLTEALGAAFDVVGAGLDRLNQ
Rv0691c|M.tuberculosis_H37Rv DFMPQTVAWTMLGVALSAYEHWLRDESVSLTEALGAAFDVVGAGLDRLNQ
MMAR_1019|M.marinum_M DLVPQTVAWTMLGVALSAYEHWLSDESVSLPEALGNAFDVVGAGLEKLD-
MUL_0771|M.ulcerans_Agy99 DLVPQTVAWTMLGVALSAYEHWLSDESVSLPEALGNAFDVVGAGLEKLD-
MAV_4481|M.avium_104 DLAPQTVAWMMLGVALSAYEHWLGDESVPLPVALGNAFDVVGAGLNGLDR
MSMEG_1420|M.smegmatis_MC2_155 DLMPQTVAWLMLGVALSAYENWLADESVSLRGAIAAAFEVARPGLDALQP
Mflv_5062|M.gilvum_PYR-GCK DLRPQTVAWTMLAVSLSAYEHWLADETACLQSSLGEAFDLLASGLSGLES
Mvan_1294|M.vanbaalenii_PYR-1 ELMPQTVAWTMLAVSLSAYEYWLADESVSLAAALGDAFDMLASGLAGLET
TH_1899|M.thermoresistible__bu DLVPQTVAWTMLGVALSAYAYWLADESVPLAQALADAFDAIRPGLDALDG
MAB_3837|M.abscessus_ATCC_1997 SLLPQTVAWTLLGVALAAYEQWLADESSKLTTLLGKGFDAAEVGLRALR-
MLBr_02568|M.leprae_Br4923 DHEYEIMAGALVGAG-EAMATRLTTGDIAVDEAAELMINLFWHGLKGAPA
. : :* ::... * * : :: **
Mb0710c|M.bovis_AF2122/97 -----------
Rv0691c|M.tuberculosis_H37Rv -----------
MMAR_1019|M.marinum_M -----------
MUL_0771|M.ulcerans_Agy99 -----------
MAV_4481|M.avium_104 -----------
MSMEG_1420|M.smegmatis_MC2_155 AH---------
Mflv_5062|M.gilvum_PYR-GCK GVSEFAPGARR
Mvan_1294|M.vanbaalenii_PYR-1 GVSEFGA----
TH_1899|M.thermoresistible__bu --KKFA-----
MAB_3837|M.abscessus_ATCC_1997 -----------
MLBr_02568|M.leprae_Br4923 DRDFGPSIVAG