For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSQAPRTPRTRYARCGDLDIAYQVIGDHPIDLLVIPGASIPVDTIDSEPSMYRFHRRLASFSRLIRFDHR GVGLSSRVSSPDALGPRFWAEDAIAVMDAAGCQQATILASGFTATTALVLAADYPERVRSLVLINASART LHAPDYELGIRSNTAEPFLTLGTDPDAVEQGFDVLRIMAPSVAHDDAFRHWWDLAGNRAASPSTARAFIN AVQASDARDSLPHITAPTLILHRVGTKFVPVEHGRYLAEHIAGSRLVELPGSDSLYWVGDTAALLDEVEE FITGVRGGFVTERVLTAVMFTGIVGSTQRAATVGDLRWRDLLDNHDNLVRHEIQRFGGREVNTAGDGFVA TFSSPSAAINCADAVVDAVAVLGIEVRVGIHAGEVEVRGADVADLAVHIGARVCALAGSSEVLVSSTVRD IVTGSSHRFAERGEHELKGVPGRWRLCALVREHLTGQR
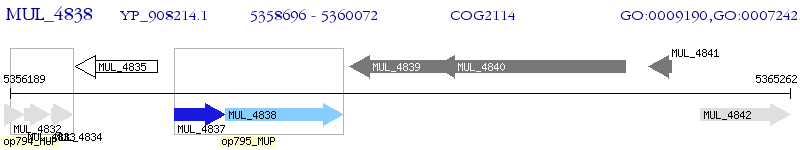
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4838 | lipJ | - | 100% (458) | lignin peroxidase LipJ |
| M. ulcerans Agy99 | MUL_0150 | - | 7e-17 | 33.33% (165) | hypothetical protein MUL_0150 |
| M. ulcerans Agy99 | MUL_2743 | - | 2e-12 | 31.08% (148) | LuxR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1935c | lipJ | 0.0 | 72.94% (462) | lignin peroxidase LIPJ |
| M. gilvum PYR-GCK | Mflv_2091 | - | 4e-17 | 35.81% (148) | putative adenylate/guanylate cyclase |
| M. tuberculosis H37Rv | Rv1900c | lipJ | 0.0 | 72.73% (462) | lignin peroxidase LIPJ |
| M. leprae Br4923 | MLBr_01899 | lipG | 1e-11 | 25.95% (289) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_3878 | - | 1e-08 | 25.00% (308) | lipase/esterase LipG |
| M. marinum M | MMAR_0286 | lipJ | 0.0 | 98.91% (458) | lignin peroxidase LipJ |
| M. avium 104 | MAV_5228 | - | 0.0 | 70.55% (455) | hydrolase, alpha/beta hydrolase fold family protein |
| M. smegmatis MC2 155 | MSMEG_4477 | - | 2e-72 | 38.26% (460) | hydrolase, alpha/beta hydrolase fold family protein |
| M. thermoresistible (build 8) | TH_1244 | - | 1e-19 | 34.97% (163) | CONSERVED HYPOTHETICAL PROTEIN |
| M. vanbaalenii PYR-1 | Mvan_4613 | - | 7e-19 | 39.33% (150) | putative adenylate/guanylate cyclase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4838|M.ulcerans_Agy99 MSQAPRTPRTRYARCGDLDIAYQVIGDHPIDLLVIPGASIPVDTIDSEPS
MMAR_0286|M.marinum_M MSQAPRTPRTRYARCGDLDIAYQVIGDHPIDLLVIPGASIPVDTIDSEPS
Mb1935c|M.bovis_AF2122/97 MAQAPHIHRTRYAKCGDMDIAYQVLGDGPTDLLVLPGPFVPIDSIDDEPS
Rv1900c|M.tuberculosis_H37Rv MAQAPHIHRTRYAKCGDMDIAYQVLGDGPTDLLVLPGPFVPIDSIDDEPS
MAV_5228|M.avium_104 MPGFSGTPRTRYASCGDTDIAYQVLGDGPIDLVLLPGPSIPIDSVDAEPS
Mflv_2091|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4613|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1244|M.thermoresistible__bu --------------------------------------------------
MSMEG_4477|M.smegmatis_MC2_155 ------MAETSYASCGDLSLAYQLFGDGPIPLVFVGPFVGHVELFWTVPE
MLBr_01899|M.leprae_Br4923 --------------------------------------------------
MAB_3878|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_4838|M.ulcerans_Agy99 MYRFHRRLASFSRLIRFDHRGVGLSSRVSSPDALGPRFWAEDAIAVMDAA
MMAR_0286|M.marinum_M MYRFHRRLASFSRLIRFDHRGVGLSSRVSSPDALGPRFWAEDAIAVMDAA
Mb1935c|M.bovis_AF2122/97 LYRFHRRLASFSRVIRLDHRGVGLSSRLAAITTLGPKFWAQDAIAVMDAV
Rv1900c|M.tuberculosis_H37Rv LYRFHRRLASFSRVIRLDHRGVGLSSRLAAITTLGPKFWAQDAIAVMDAV
MAV_5228|M.avium_104 MYRFHRRLAAFARVIRLDQRGIGLSSRVPSVDVIGPEFWARDVVSVMDAV
Mflv_2091|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4613|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1244|M.thermoresistible__bu --------------------------------VAARPLPLAVAAAPGSPI
MSMEG_4477|M.smegmatis_MC2_155 FKSFFDQLASFCRVAIFDKAGTGLSDPVPKVRTLDDR--AAEIEAVMDAV
MLBr_01899|M.leprae_Br4923 ------------------------------MEVRSGYATSGDLKLYYEDM
MAB_3878|M.abscessus_ATCC_1997 -----------------------MVPLSSEVPTRSGIARNGDVELFYEDL
MUL_4838|M.ulcerans_Agy99 GCQQATILASGFTATTALVLAADYPERVRSLVLINASARTL---------
MMAR_0286|M.marinum_M GCQEATILASGFTATTALVLAADYPERVRSLVLINASARTL---------
Mb1935c|M.bovis_AF2122/97 GCEQATIFAPSFHAMNGLVLAADYPERVRSLIVVNGSARPL---------
Rv1900c|M.tuberculosis_H37Rv GCEQATIFAPSFHAMNGLVLAADYPERVRSLIVVNGSARPL---------
MAV_5228|M.avium_104 GCERATVVAPSFTSMTGLVLAADYPDRVSSLVIINGAARTV---------
Mflv_2091|M.gilvum_PYR-GCK ------------MLILVCVLAVLAAGLGIALAVQTRRLTSA---------
Mvan_4613|M.vanbaalenii_PYR-1 --------------MALLVLAAVVVGLAVALAVQSRRLRTA---------
TH_1244|M.thermoresistible__bu PRRVGQDGAVTQIAIVVVALAGLALAEAVALAVLWGRLRRE---------
MSMEG_4477|M.smegmatis_MC2_155 GFDDAVVLAMSEGGPASILFAANRPGRVRTLVAYGSFATMAGCRWEDLDG
MLBr_01899|M.leprae_Br4923 GNVDDPPVLLIMGLGAQLVLWR--TAFCEKLVAQGLRVVRYD--------
MAB_3878|M.abscessus_ATCC_1997 GNPGDPAVVLVMGVAAQLPMWP--DGFCRQLLDQGYRVIRFD--------
: *
MUL_4838|M.ulcerans_Agy99 ------HAPDYELGIRSNTAEPFLTLGTDPDAVEQGFDVLRIMAPSVAHD
MMAR_0286|M.marinum_M ------HAPDYELGIRSNTAEPFLTLGTDPDAVEQGFDVLRIMAPSVAHD
Mb1935c|M.bovis_AF2122/97 ------WAPDYPVGAQVRRADPFLTVALEPDAVEQGFDVLSIVAPTVAGD
Rv1900c|M.tuberculosis_H37Rv ------WAPDYPVGAQVRRADPFLTVALEPDAVERGFDVLSIVAPTVAGD
MAV_5228|M.avium_104 ------YAPDYPMGARIETLDPYTTVAVDPDAVDQGFDMLGLIAPSVAGD
Mflv_2091|M.gilvum_PYR-GCK ------RQEAEELRHRLDTRQLLVSGGTK---------------------
Mvan_4613|M.vanbaalenii_PYR-1 ------RQEADELRRRLDTRQMLVTGSTK---------------------
TH_1244|M.thermoresistible__bu ------RRAADELRRRLDTRTTLMAGGRE---------------------
MSMEG_4477|M.smegmatis_MC2_155 DPVDIRARSVAEMGDEYAPTVEQVIHFQEQARAVRGQWGSGAAVRCAMPS
MLBr_01899|M.leprae_Br4923 -------NRDVGLSSRTDSTEQPCPSQPLTAR------------------
MAB_3878|M.abscessus_ATCC_1997 -------NRDCGLSTKLDGQKAPG---SVQRR------------------
: .
MUL_4838|M.ulcerans_Agy99 DAFRHWWDLAGNRAASPSTARAFINAVQASDARDSLPHITAPTLILHRVG
MMAR_0286|M.marinum_M DAFRDWWDLAGNRAASPSTARAFINAVQASDARDSLPHITAPTLILHRVG
Mb1935c|M.bovis_AF2122/97 DVFRAWWDLAGNRAGPPSMARAVSKVIAEADVRDVLGHIEAPTLILHRVG
Rv1900c|M.tuberculosis_H37Rv DVFRAWWDLAGNRAGPPSIARAVSKVIAEADVRDVLGHIEAPTLILHRVG
MAV_5228|M.avium_104 AAFRSWWDAAGNRAASPSMARAMILSIRHADVRDRLARITTPTLILHRTD
Mflv_2091|M.gilvum_PYR-GCK -AVKTVWQTAN---------------------------------ILRKDG
Mvan_4613|M.vanbaalenii_PYR-1 -AVKTVWQTAN---------------------------------ILRRDG
TH_1244|M.thermoresistible__bu -AVRTVWQTAN---------------------------------ILRTDG
MSMEG_4477|M.smegmatis_MC2_155 MGSMRQLGMFERLCASPGMARATFEAAFRIDVRPILPTIAAPTLVIHARD
MLBr_01899|M.leprae_Br4923 -LIRFWLGQRN----------------------------ACAYTLEDMTD
MAB_3878|M.abscessus_ATCC_1997 -VVRYALGMGS----------------------------KVPYTLIDMAE
:
MUL_4838|M.ulcerans_Agy99 TKFVPVEHGRYLAEHIAGSRLVELPGSDSLYWVGDTAALLDEVEEFITGV
MMAR_0286|M.marinum_M TKFVPVEHGRYLAEHIAGSRLVELPGSDSLYWVGDTAALLDEVEEFITGV
Mb1935c|M.bovis_AF2122/97 STYIPVGHGRYLAEHIAGSRLVELPGTDTLYWVGDTGPMLDEIEEFITGV
Rv1900c|M.tuberculosis_H37Rv STYIPVGHGRYLAEHIAGSRLVELPGTDTLYWVGDTGPMLDEIEEFITGV
MAV_5228|M.avium_104 CVFTSVDHGRYLAEHIAGSRFVSLPGEDTLYWVGDTGPMLDEIEEFITGV
Mflv_2091|M.gilvum_PYR-GCK FGAAVRSS----IEELADWAEVERPDLARLSLNGR---------------
Mvan_4613|M.vanbaalenii_PYR-1 FGAAVRSS----IEELADWAEVERPDLARLAPNGR---------------
TH_1244|M.thermoresistible__bu LGAAARSS----IEDLADWAEVERPDLARLAANGK---------------
MSMEG_4477|M.smegmatis_MC2_155 DPAVPVQCSRYLADHISGARWLEVDGVDHAPWFTEADRVLTEIEEFVTGN
MLBr_01899|M.leprae_Br4923 DAVALLDHLSIERAHIVGASMGGMIAQIFAARFPTR--------------
MAB_3878|M.abscessus_ATCC_1997 DVRSLVDHLGLEKVHIAGASMGGMIVQVFAGTYPER--------------
.: .
MUL_4838|M.ulcerans_Agy99 RGGFV-TERVLTAVMFTGIVGSTQRAATVGDLRWRDLLDNHDNLVRHEIQ
MMAR_0286|M.marinum_M RGGFV-TERVLTTVMFTDIVGSTQRAATVGDLRWRDLLDNHDNLVRHEIQ
Mb1935c|M.bovis_AF2122/97 RGGAD-AERMLATIMFTDIVGSTQHAAALGDDRWRDLLDNHDTIVCHEIQ
Rv1900c|M.tuberculosis_H37Rv RGGAD-AERMLATIMFTDIVGSTQHAAALGDDRWRDLLDNHDTIVCHEIQ
MAV_5228|M.avium_104 RGGSE-AERLLTTIVFTDIVGSTERAAALGDYRWRDLLDNHDRIVRHELQ
Mflv_2091|M.gilvum_PYR-GCK -----------LTIMFSDIEESTALNERIGDRAFVRLIGRHDKSVRKFVD
Mvan_4613|M.vanbaalenii_PYR-1 -----------VAIMFSDIEESTALNERIGDRAFVRLLGRHDKPVRRHVD
TH_1244|M.thermoresistible__bu -----------VVILFSDIEGSTALNNRIGDRAWVRLIDRHSRLVRRFVK
MSMEG_4477|M.smegmatis_MC2_155 HAAPHNSHRALRTVLFTDIVASTQHAASNGDERWRAVLQRFGEVTEELSG
MLBr_01899|M.leprae_Br4923 ----------TRSLAVFFSSNNRPFLPPPAPRALLALLTGPPTGSRRDVV
MAB_3878|M.abscessus_ATCC_1997 ----------VHSVGIIYSATGRPFSRLPSWELIKTALNAPGKNATAEEW
: . . . :
MUL_4838|M.ulcerans_Agy99 RFGGREVNTAGDGFVATFSSPSAAINCADAVVDAVAVLG--IEVRVGIHA
MMAR_0286|M.marinum_M RFGGREVNTAGDGFVATFSSPSAAINCADAVVDAVAVLG--IEVRVGIHA
Mb1935c|M.bovis_AF2122/97 RFGGREVNTAGDGFVATFTSPSAAIACADDIVDAVAALG--IEVRIGIHA
Rv1900c|M.tuberculosis_H37Rv RFGGREVNTAGDGFVATFTSPSAAIACADDIVDAVAALG--IEVRIGIHA
MAV_5228|M.avium_104 RFGGREVNTAGDGFVATFSSPSAAISCADALVDAVAALG--IEVRVGIHA
Mflv_2091|M.gilvum_PYR-GCK KHEGHVVKSQGDGFMVAFAKPEQAVRCGLDIQHSLARRPNDIRVRMGVHT
Mvan_4613|M.vanbaalenii_PYR-1 AHDGYVVKSQGDGFMVAFARPEQAVRCGLAIQHSLDKRPNGIRVRIGIHV
TH_1244|M.thermoresistible__bu KHDGYVIKSQGDGFMVAFARPEQAVRCSVELQRALRKRPDRIRVRIGIHL
MSMEG_4477|M.smegmatis_MC2_155 RFGGVVVKSTGDGHLSTFEGPTHAIRYAEALRSDAETLG--IQIRAAIHT
MLBr_01899|M.leprae_Br4923 VDNVVRVTKITGSPLYRMPEEQVRTNAAEIYDRSFYPLGVSRQFSAILGS
MAB_3878|M.abscessus_ATCC_1997 LEFEVNNGIVYNGPDNLPSREQLRQRILDHRARSDYKIGTVRQFDAILGT
.. .. :
MUL_4838|M.ulcerans_Agy99 GEVEVR----GADVADLAVHIGARVCALAGSSEVLVSSTVRDIVTGSSHR
MMAR_0286|M.marinum_M GEVEVR----GADVAGLAVHIGARVCALAGSSEVLVSSTVRDIVTGSSHR
Mb1935c|M.bovis_AF2122/97 GEVEVRDASHGTDVAGVAVHIGARVCALAGPSEVLVSSTVRDIVAGSRHR
Rv1900c|M.tuberculosis_H37Rv GEVEVRDASHGTDVAGVAVHIGARVCALAGPSEVLVSSTVRDIVAGSRHR
MAV_5228|M.avium_104 GEVEVRG-ARKDDIAGMAVHIGARVAALAGPGEVLVSSTLRDIVTGSRHR
Mflv_2091|M.gilvum_PYR-GCK GKSVLR----GDDVFGRNVAMAARVAAAADGGEILVSSAVHRALADCEDL
Mvan_4613|M.vanbaalenii_PYR-1 GKSVLR----GDDLFGRNVAMAARVAAEADGGEVLVSSAVRDALTDADDI
TH_1244|M.thermoresistible__bu GRSVRR----GDDLFGRNVALAARVAGAAEGGEVLVSQAVRDALNSAGDA
MSMEG_4477|M.smegmatis_MC2_155 GECELLG----DDIGGIAVHIAARILGQARAGEILVSRTVRDLVVGSGTS
MLBr_01899|M.leprae_Br4923 GSLLHYN----QRIIAPTVVIHGRADKLVRPS---SGRAVARAITGARLV
MAB_3878|M.abscessus_ATCC_1997 GSLLRFT----RAVVAPAVVIHGRNDPLVPYQ---NGRVVAKNMRNARFA
* : * : .* . . .: : ..
MUL_4838|M.ulcerans_Agy99 FA---------ERGEHELKGVPGRWRLCALVREHLTGQR-----------
MMAR_0286|M.marinum_M FA---------ERGEHELKGVPGRWRLCALVREHLTGQR-----------
Mb1935c|M.bovis_AF2122/97 FA---------ERGEQELKGVPGRWRLCVLMRDDATRTR-----------
Rv1900c|M.tuberculosis_H37Rv FA---------ERGEQELKGVPGRWRLCVLMRDDATRTR-----------
MAV_5228|M.avium_104 FG---------DRGETALKGVPGQWRLYRLLREHAAAER-----------
Mflv_2091|M.gilvum_PYR-GCK A--------FDEQRDVELKGFSGSHTLYPVRSASASR-------------
Mvan_4613|M.vanbaalenii_PYR-1 V--------VDGERDAHLKGFSGSHTLYSVRSASASR-------------
TH_1244|M.thermoresistible__bu GSDEVAGFDVDDGREVSLKGFPGRHRLFAVRGQAGFRPRPRPE-------
MSMEG_4477|M.smegmatis_MC2_155 FA---------DRGNVELRGVPGTWQLLAVGDGAGGGSPAGSVEALLAAE
MLBr_01899|M.leprae_Br4923 LF---------DGMGHDLPQQLWDQAVGVLMSNFAKAS------------
MAB_3878|M.abscessus_ATCC_1997 LI---------ERMGHDLPEPVWKPVTSELLTTFECAVQD----------
* :
MUL_4838|M.ulcerans_Agy99 ----------------------------------------
MMAR_0286|M.marinum_M ----------------------------------------
Mb1935c|M.bovis_AF2122/97 ----------------------------------------
Rv1900c|M.tuberculosis_H37Rv ----------------------------------------
MAV_5228|M.avium_104 ----------------------------------------
Mflv_2091|M.gilvum_PYR-GCK ----------------------------------------
Mvan_4613|M.vanbaalenii_PYR-1 ----------------------------------------
TH_1244|M.thermoresistible__bu ----------------------------------------
MSMEG_4477|M.smegmatis_MC2_155 PTPAARTATRRADRAVAAVAKRTPWALRGVARLTMSATRR
MLBr_01899|M.leprae_Br4923 ----------------------------------------
MAB_3878|M.abscessus_ATCC_1997 ----------------------------------------