For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AETSYASCGDLSLAYQLFGDGPIPLVFVGPFVGHVELFWTVPEFKSFFDQLASFCRVAIFDKAGTGLSDP VPKVRTLDDRAAEIEAVMDAVGFDDAVVLAMSEGGPASILFAANRPGRVRTLVAYGSFATMAGCRWEDLD GDPVDIRARSVAEMGDEYAPTVEQVIHFQEQARAVRGQWGSGAAVRCAMPSMGSMRQLGMFERLCASPGM ARATFEAAFRIDVRPILPTIAAPTLVIHARDDPAVPVQCSRYLADHISGARWLEVDGVDHAPWFTEADRV LTEIEEFVTGNHAAPHNSHRALRTVLFTDIVASTQHAASNGDERWRAVLQRFGEVTEELSGRFGGVVVKS TGDGHLSTFEGPTHAIRYAEALRSDAETLGIQIRAAIHTGECELLGDDIGGIAVHIAARILGQARAGEIL VSRTVRDLVVGSGTSFADRGNVELRGVPGTWQLLAVGDGAGGGSPAGSVEALLAAEPTPAARTATRRADR AVAAVAKRTPWALRGVARLTMSATRR*
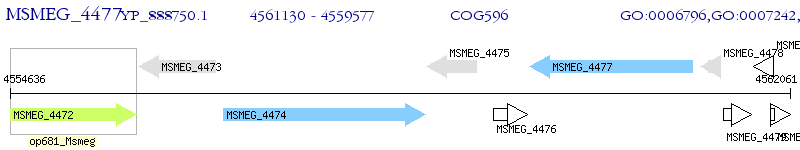
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4477 | - | - | 100% (517) | hydrolase, alpha/beta hydrolase fold family protein |
| M. smegmatis MC2 155 | MSMEG_3856 | - | 1e-21 | 29.60% (277) | transcriptional regulator, CadC |
| M. smegmatis MC2 155 | MSMEG_3297 | - | 9e-18 | 28.16% (277) | transcriptional regulator, CadC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1935c | lipJ | 2e-73 | 37.82% (468) | lignin peroxidase LIPJ |
| M. gilvum PYR-GCK | Mflv_3121 | - | 1e-21 | 29.24% (277) | transcriptional regulator, CadC |
| M. tuberculosis H37Rv | Rv1900c | lipJ | 6e-73 | 37.61% (468) | lignin peroxidase LIPJ |
| M. leprae Br4923 | MLBr_01399 | - | 5e-13 | 32.06% (131) | hypothetical protein MLBr_01399 |
| M. abscessus ATCC 19977 | MAB_2330 | - | 1e-12 | 34.40% (125) | putative purine cyclase-like protein |
| M. marinum M | MMAR_0286 | lipJ | 1e-74 | 38.91% (460) | lignin peroxidase LipJ |
| M. avium 104 | MAV_5228 | - | 1e-73 | 38.79% (464) | hydrolase, alpha/beta hydrolase fold family protein |
| M. thermoresistible (build 8) | TH_1244 | - | 5e-19 | 34.71% (170) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4838 | lipJ | 1e-72 | 38.26% (460) | lignin peroxidase LipJ |
| M. vanbaalenii PYR-1 | Mvan_3415 | - | 5e-18 | 27.41% (259) | transcriptional regulator, CadC |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1935c|M.bovis_AF2122/97 MAQAPHIHRTRYAKCGDMDIAYQVLGDGPTDLLVLPGPFVPIDSIDDEPS
Rv1900c|M.tuberculosis_H37Rv MAQAPHIHRTRYAKCGDMDIAYQVLGDGPTDLLVLPGPFVPIDSIDDEPS
MMAR_0286|M.marinum_M MSQAPRTPRTRYARCGDLDIAYQVIGDHPIDLLVIPGASIPVDTIDSEPS
MUL_4838|M.ulcerans_Agy99 MSQAPRTPRTRYARCGDLDIAYQVIGDHPIDLLVIPGASIPVDTIDSEPS
MAV_5228|M.avium_104 MPGFSGTPRTRYASCGDTDIAYQVLGDGPIDLVLLPGPSIPIDSVDAEPS
MSMEG_4477|M.smegmatis_MC2_155 ------MAETSYASCGDLSLAYQLFGDGPIPLVFVGPFVGHVELFWTVPE
MLBr_01399|M.leprae_Br4923 --------------------------------------------------
MAB_2330|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3121|M.gilvum_PYR-GCK -------------------------------MTSAD-ALSSSPRVWRWDD
Mvan_3415|M.vanbaalenii_PYR-1 ---------MPPTAGADLAATRGCRHRYCCRVTSADSALSPHRRVWRWDD
TH_1244|M.thermoresistible__bu --------------------------------------------------
Mb1935c|M.bovis_AF2122/97 LYRFHRRLASFSRVIRLDHRGVGLSSRLAAITTLGPKFWAQDAIAVMDAV
Rv1900c|M.tuberculosis_H37Rv LYRFHRRLASFSRVIRLDHRGVGLSSRLAAITTLGPKFWAQDAIAVMDAV
MMAR_0286|M.marinum_M MYRFHRRLASFSRLIRFDHRGVGLSSRVSSPDALGPRFWAEDAIAVMDAA
MUL_4838|M.ulcerans_Agy99 MYRFHRRLASFSRLIRFDHRGVGLSSRVSSPDALGPRFWAEDAIAVMDAA
MAV_5228|M.avium_104 MYRFHRRLAAFARVIRLDQRGIGLSSRVPSVDVIGPEFWARDVVSVMDAV
MSMEG_4477|M.smegmatis_MC2_155 FKSFFDQLASFCRVAIFDKAGTGLSDPVPKVRTLDDR--AAEIEAVMDAV
MLBr_01399|M.leprae_Br4923 --------------------------------------------------
MAB_2330|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_3121|M.gilvum_PYR-GCK YVLDLGRYELRAGDSVIRVEPQVFDVLTQLVSHHDRCVSKEELFDTVWGG
Mvan_3415|M.vanbaalenii_PYR-1 FVLDVGCYELRRGDTMIRVEPQVFDVLAQLVSNHGRFVSKEELFDTVWGG
TH_1244|M.thermoresistible__bu --------------------------------------------------
Mb1935c|M.bovis_AF2122/97 GCEQATIFAPSFHAMNGLVLAADYPERVRSLIVVN-----GSARPLWAPD
Rv1900c|M.tuberculosis_H37Rv GCEQATIFAPSFHAMNGLVLAADYPERVRSLIVVN-----GSARPLWAPD
MMAR_0286|M.marinum_M GCQEATILASGFTATTALVLAADYPERVRSLVLIN-----ASARTLHAPD
MUL_4838|M.ulcerans_Agy99 GCQQATILASGFTATTALVLAADYPERVRSLVLIN-----ASARTLHAPD
MAV_5228|M.avium_104 GCERATVVAPSFTSMTGLVLAADYPDRVSSLVIIN-----GAARTVYAPD
MSMEG_4477|M.smegmatis_MC2_155 GFDDAVVLAMSEGGPASILFAANRPGRVRTLVAYGSFATMAGCRWEDLDG
MLBr_01399|M.leprae_Br4923 --MPPSAKSTYP-------------GQVEGAPHDG---------------
MAB_2330|M.abscessus_ATCC_1997 MDVEPSGLDAEPDGEFDSGIRKRRRGRTRGPVRHD---------------
Mflv_3121|M.gilvum_PYR-GCK RFVGEAALTSRIKAVRRALGDDGEAQRYIRTVRGR---------------
Mvan_3415|M.vanbaalenii_PYR-1 RFVGEAALTSRIKAVRRALGDDGEAQRYIRTVRGR---------------
TH_1244|M.thermoresistible__bu --VAARPLPLAVAAAPG---------------------------------
Mb1935c|M.bovis_AF2122/97 YPVGAQVR-----------RADPFLTVALEPDAVEQGFDVLSIVAPTVAG
Rv1900c|M.tuberculosis_H37Rv YPVGAQVR-----------RADPFLTVALEPDAVERGFDVLSIVAPTVAG
MMAR_0286|M.marinum_M YELGIRSN-----------TAEPFLTLGTDPDAVEQGFDVLRIMAPSVAH
MUL_4838|M.ulcerans_Agy99 YELGIRSN-----------TAEPFLTLGTDPDAVEQGFDVLRIMAPSVAH
MAV_5228|M.avium_104 YPMGARIE-----------TLDPYTTVAVDPDAVDQGFDMLGLIAPSVAG
MSMEG_4477|M.smegmatis_MC2_155 DPVDIRARSVAEMGDEYAPTVEQVIHFQEQARAVRGQWGSGAAVRCAMPS
MLBr_01399|M.leprae_Br4923 ---------------------TPSAPQEPDEDTVTVPAPALIRRSSVSMP
MAB_2330|M.abscessus_ATCC_1997 ---------------------SAQVTDGIDTTADEVPSTESASRHFARAS
Mflv_3121|M.gilvum_PYR-GCK -------------------GYRFVGTLHGEDSATPDEASVEHV---VRET
Mvan_3415|M.vanbaalenii_PYR-1 -------------------GYQFVGRLLDDGTAPAPTAAPEPAPEPVPEP
TH_1244|M.thermoresistible__bu --------------------SPIPRRVGQDGAVTQIAIVVVALAGLALAE
: .
Mb1935c|M.bovis_AF2122/97 DDVFRAWWDLAGNRAGPPSMARAVSKVIAEADVRDVLGHIEAPTLILHRV
Rv1900c|M.tuberculosis_H37Rv DDVFRAWWDLAGNRAGPPSIARAVSKVIAEADVRDVLGHIEAPTLILHRV
MMAR_0286|M.marinum_M DDAFRDWWDLAGNRAASPSTARAFINAVQASDARDSLPHITAPTLILHRV
MUL_4838|M.ulcerans_Agy99 DDAFRHWWDLAGNRAASPSTARAFINAVQASDARDSLPHITAPTLILHRV
MAV_5228|M.avium_104 DAAFRSWWDAAGNRAASPSMARAMILSIRHADVRDRLARITTPTLILHRT
MSMEG_4477|M.smegmatis_MC2_155 MGSMRQLG-MFERLCASPGMARATFEAAFRIDVRPILPTIAAPTLVIHAR
MLBr_01399|M.leprae_Br4923 -NAAQWLHTTNRSPRLVAMVRRARRLLPGDPDFGDPLSTAGE-------G
MAB_2330|M.abscessus_ATCC_1997 RTAGSWFRKMDRDPAVVAALRRARRSLPGDPYFGDPLSTSGF-------G
Mflv_3121|M.gilvum_PYR-GCK PRQHIAFARAADGVRLAYAESGDGPPLVKAANWMTHLGYDSESPVWRHGV
Mvan_3415|M.vanbaalenii_PYR-1 PRQHIAFTRAADGVRLAYAVSGEGRPLVRAANWMTHLGYDIESPVWRHWV
TH_1244|M.thermoresistible__bu AVALAVLWGRLRRE------RRAADELRRRLDTRTTLMAGGR-------E
*
Mb1935c|M.bovis_AF2122/97 GSTYIPVGHGRYLAEHIAGSRLVELPGTDTLYWVGDTGPMLDEIEEFITG
Rv1900c|M.tuberculosis_H37Rv GSTYIPVGHGRYLAEHIAGSRLVELPGTDTLYWVGDTGPMLDEIEEFITG
MMAR_0286|M.marinum_M GTKFVPVEHGRYLAEHIAGSRLVELPGSDSLYWVGDTAALLDEVEEFITG
MUL_4838|M.ulcerans_Agy99 GTKFVPVEHGRYLAEHIAGSRLVELPGSDSLYWVGDTAALLDEVEEFITG
MAV_5228|M.avium_104 DCVFTSVDHGRYLAEHIAGSRFVSLPGEDTLYWVGDTGPMLDEIEEFITG
MSMEG_4477|M.smegmatis_MC2_155 DDPAVPVQCSRYLADHISGARWLEVDGVDHAPWFTEADRVLTEIEEFVTG
MLBr_01399|M.leprae_Br4923 GPRAAARAADRLLGDRGAASREVSLS-------------VLQVWQALTEA
MAB_2330|M.abscessus_ATCC_1997 GASAVARAADRLVTDRDTAVREFGFG-------------ALQIWQAFTEK
Mflv_3121|M.gilvum_PYR-GCK REMSLRYRFIRYD-ERGCGLSDWDVGDFTFDDWVADLETVVDAVGLDRFH
Mvan_3415|M.vanbaalenii_PYR-1 RDMSLRHKFIRYD-ERGCGLSDWEVGAFTFDDWVTDLESVVEAVGLERFP
TH_1244|M.thermoresistible__bu AVRTVWQTANILRTDGLGAAARSSIED-------------LADWAEVERP
: . . :
Mb1935c|M.bovis_AF2122/97 VRGG-ADAERMLATIMFTDIVGSTQHAAALGDDRWRDLLDNHDTIVCHEI
Rv1900c|M.tuberculosis_H37Rv VRGG-ADAERMLATIMFTDIVGSTQHAAALGDDRWRDLLDNHDTIVCHEI
MMAR_0286|M.marinum_M VRGG-FVTERVLTTVMFTDIVGSTQRAATVGDLRWRDLLDNHDNLVRHEI
MUL_4838|M.ulcerans_Agy99 VRGG-FVTERVLTAVMFTGIVGSTQRAATVGDLRWRDLLDNHDNLVRHEI
MAV_5228|M.avium_104 VRGG-SEAERLLTTIVFTDIVGSTERAAALGDYRWRDLLDNHDRIVRHEL
MSMEG_4477|M.smegmatis_MC2_155 NHAAPHNSHRALRTVLFTDIVASTQHAASNGDERWRAVLQRFGEVTEELS
MLBr_01399|M.leprae_Br4923 IARR---PVNPEVTLVFTDLVGFSGWSLQAGDEATLALLRQVARAVESPL
MAB_2330|M.abscessus_ATCC_1997 VSKQ---PANREVTLVFTDLVGFSGWALEAGDEATLKLLRRVASVSEPPL
Mflv_3121|M.gilvum_PYR-GCK LMGV---SQGGAVAVAYAARHPDKVSRLVLCGAYARGRAVRALSDDEKRA
Mvan_3415|M.vanbaalenii_PYR-1 LLGV---SQGGAVAVAYAARHPDRVTRLVLCGAYARGRVVRAMNDDEKRA
TH_1244|M.thermoresistible__bu DLAR--LAANGKVVILFSDIEGSTALNNRIGDRAWVRLIDRHSRLVRRFV
. .: :: . .
Mb1935c|M.bovis_AF2122/97 QRFGGREVNTAGDGFVATFTSPSAAIACADDIVDAVAALGIE-----VRI
Rv1900c|M.tuberculosis_H37Rv QRFGGREVNTAGDGFVATFTSPSAAIACADDIVDAVAALGIE-----VRI
MMAR_0286|M.marinum_M QRFGGREVNTAGDGFVATFSSPSAAINCADAVVDAVAVLGIE-----VRV
MUL_4838|M.ulcerans_Agy99 QRFGGREVNTAGDGFVATFSSPSAAINCADAVVDAVAVLGIE-----VRV
MAV_5228|M.avium_104 QRFGGREVNTAGDGFVATFSSPSAAISCADALVDAVAALGIE-----VRV
MSMEG_4477|M.smegmatis_MC2_155 GRFGGVVVKSTGDGHLSTFEGPTHAIRYAEALRSDAETLGIQ-----IRA
MLBr_01399|M.leprae_Br4923 LDAGGHIVKRMGDGIMAVFRDPSVAVQAVLAATEAMKSVEVGGYTPRIRV
MAB_2330|M.abscessus_ATCC_1997 LSAGGQVVKRMGDGIMAVFDDPVTALRAIIPARDALKNVEIQGYTPVMRI
Mflv_3121|M.gilvum_PYR-GCK AALDLELARVGWGRDDPAFRQVFAAQFLPDGTRADWAAFDQLQR----RT
Mvan_3415|M.vanbaalenii_PYR-1 AALDLDLARVGWGRDDPAFRQVFAAQFLPDGTRADWAAFDQLQR----RT
TH_1244|M.thermoresistible__bu KKHDGYVIKSQGDGFMVAFARPEQAVRCSVELQRALRKRPDRIR---VRI
. . . .* * *
Mb1935c|M.bovis_AF2122/97 GIHAGEVEVRDASHGTDVAGVAVHIGARVCALAGPSEVLVSS----TVRD
Rv1900c|M.tuberculosis_H37Rv GIHAGEVEVRDASHGTDVAGVAVHIGARVCALAGPSEVLVSS----TVRD
MMAR_0286|M.marinum_M GIHAGEVEVR----GADVAGLAVHIGARVCALAGSSEVLVSS----TVRD
MUL_4838|M.ulcerans_Agy99 GIHAGEVEVR----GADVADLAVHIGARVCALAGSSEVLVSS----TVRD
MAV_5228|M.avium_104 GIHAGEVEVRG-ARKDDIAGMAVHIGARVAALAGPGEVLVSS----TLRD
MSMEG_4477|M.smegmatis_MC2_155 AIHTGECELLG----DDIGGIAVHIAARILGQARAGEILVSR----TVRD
MLBr_01399|M.leprae_Br4923 GIHTGRPQRLA----ADWLGVDVNIAARVMERATKGGIMISG----PTLD
MAB_2330|M.abscessus_ATCC_1997 GIHAGTPQRIG----SDWLGVDVNIAARVMESAAKGGLAISQ----PALA
Mflv_3121|M.gilvum_PYR-GCK TSPENAVHFLEEFARIDVRDLCARVQCPTLVLHSRDDHRVPMRYGEELAS
Mvan_3415|M.vanbaalenii_PYR-1 TSPENAVRFLEEFAHIDVRDQCGQVRCPTLILHSRDDHRVPMRYGEELAA
TH_1244|M.thermoresistible__bu GIHLGRSVRRG----DDLFGRNVALAARVAGAAEGGEVLVSQAVRDALNS
. * . : . . :.
Mb1935c|M.bovis_AF2122/97 IVAGSRHRFAERGEQELK---------GVPGRWRLCVL------------
Rv1900c|M.tuberculosis_H37Rv IVAGSRHRFAERGEQELK---------GVPGRWRLCVL------------
MMAR_0286|M.marinum_M IVTGSSHRFAERGEHELK---------GVPGRWRLCAL------------
MUL_4838|M.ulcerans_Agy99 IVTGSSHRFAERGEHELK---------GVPGRWRLCAL------------
MAV_5228|M.avium_104 IVTGSRHRFGDRGETALK---------GVPGQWRLYRL------------
MSMEG_4477|M.smegmatis_MC2_155 LVVGSGTSFADRGNVELR---------GVPGTWQLLAVGDGAGGGSPAGS
MLBr_01399|M.leprae_Br4923 LIPQSDLKELGIITRRVRKPMFTSKFTGIPPDMVIYRIK-----------
MAB_2330|M.abscessus_ATCC_1997 KVPLAFLDHMGLTPTPVRRPLFSSRPAGVPENMKMYLLE-----------
Mflv_3121|M.gilvum_PYR-GCK LIPDARLVALSSNNHLLT---------ATEPAWQVFCA------------
Mvan_3415|M.vanbaalenii_PYR-1 LIPDARLVALSSNNHLLT---------GAEPAWRVFCD------------
TH_1244|M.thermoresistible__bu AGDAGSDEVAGFDVDDGRE----VSLKGFPGRHRLFAVR-----------
. . :
Mb1935c|M.bovis_AF2122/97 --------------------------------------MRDDATRTR---
Rv1900c|M.tuberculosis_H37Rv --------------------------------------MRDDATRTR---
MMAR_0286|M.marinum_M --------------------------------------VREHLTGQR---
MUL_4838|M.ulcerans_Agy99 --------------------------------------VREHLTGQR---
MAV_5228|M.avium_104 --------------------------------------LREHAAAER---
MSMEG_4477|M.smegmatis_MC2_155 VEALLAAEPTPAARTATRRADRAVAAVAKRTPWALRGVARLTMSATRR--
MLBr_01399|M.leprae_Br4923 --------------------------------------ARRELTASDETA
MAB_2330|M.abscessus_ATCC_1997 --------------------------------------TRRQLPAPGDDG
Mflv_3121|M.gilvum_PYR-GCK --------------------------------------EVAAFLV-----
Mvan_3415|M.vanbaalenii_PYR-1 --------------------------------------EVETFLAV----
TH_1244|M.thermoresistible__bu --------------------------------------GQAGFRPRPRPE
Mb1935c|M.bovis_AF2122/97 ------
Rv1900c|M.tuberculosis_H37Rv ------
MMAR_0286|M.marinum_M ------
MUL_4838|M.ulcerans_Agy99 ------
MAV_5228|M.avium_104 ------
MSMEG_4477|M.smegmatis_MC2_155 ------
MLBr_01399|M.leprae_Br4923 QTNSLT
MAB_2330|M.abscessus_ATCC_1997 RV----
Mflv_3121|M.gilvum_PYR-GCK ------
Mvan_3415|M.vanbaalenii_PYR-1 ------
TH_1244|M.thermoresistible__bu ------