For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQVAIITAATGGIGFGCAAKFAEMSMAVVGTARDERRLAELTQLPGGPDRIATHPVDPTDDDAPGQLTRA ALDRWGRIDFLINAAGEGSPKSVHETDDESLDYVLSLMLRAPFRLTREVLPHLRPGSAIINITSTFAVVG GLRGGAYSAAKGGLAALTTHIACQYGAQGIRCNAVAPGVIQTAMTQHRLADERFRRLNVEITPHQRLGTV DDVANTVAFLCSEGGAFINGQTIVVDGG
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
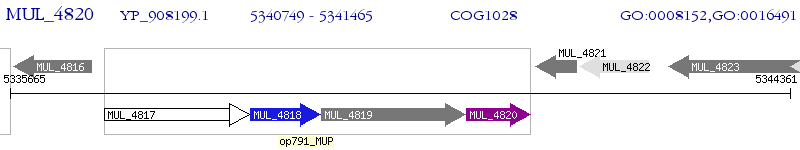
| M. ulcerans Agy99 | MUL_4820 | - | - | 100% (238) | short-chain type dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_4361 | - | 4e-28 | 33.47% (239) | short-chain type dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_4442 | - | 7e-25 | 33.47% (242) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 4e-25 | 33.47% (245) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1783 | - | 3e-97 | 72.15% (237) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2857c | - | 4e-25 | 33.47% (245) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 8e-21 | 28.99% (238) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4632 | - | 2e-27 | 33.05% (239) | short chain dehydrogenase |
| M. marinum M | MMAR_0302 | - | 1e-129 | 94.96% (238) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_5213 | - | 1e-100 | 74.37% (238) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_5632 | - | 1e-101 | 74.79% (238) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_4328 | - | 2e-94 | 72.31% (242) | PUTATIVE oxidoreductase, short chain dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_4967 | - | 2e-97 | 73.95% (238) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4820|M.ulcerans_Agy99 --------------------MQVAIITAATGGIGFGCAAKFAEMSMAVVG
MMAR_0302|M.marinum_M --------------------MQVAIITGATGGIGFGCAAKLAEMGMAVLG
MSMEG_5632|M.smegmatis_MC2_155 --------------------MQVAIITGASSGIGFGCATKLAEAGMAVLG
TH_4328|M.thermoresistible__bu ------------------VNMQVAIVTGASSGIGFGCATKLAEQGMAVLG
Mflv_1783|M.gilvum_PYR-GCK -----------------MQTAQVAIVTGASSGIGFGCATALADRGMVVLG
Mvan_4967|M.vanbaalenii_PYR-1 --------------------MQVAIVTGASSGIGFGCATTLAEQGMAVLG
MAV_5213|M.avium_104 --------------------MQVAIVTGASSGIGLGCATRLAGTGMAVLG
MAB_4632|M.abscessus_ATCC_1997 -----------MILDRFKLDGNVAVVTGAGRGLGAAIAEAFAQAGADVLI
Mb2882c|M.bovis_AF2122/97 MMDLSQRLAG-----------RVAVITGGGSGIGLAAGRRMRAEGATIVV
Rv2857c|M.tuberculosis_H37Rv MMDLSQRLAG-----------RVAVITGGGSGIGLAAGRRMRAEGATIVV
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
. ::*.. *:* . . : . :
MUL_4820|M.ulcerans_Agy99 TARDERRLAELTQLPGGP----DRIATHPVDPTDDDAPGQLTRAALDRWG
MMAR_0302|M.marinum_M TGRDERRLAELTQLPGGP----DRIATHPVDLTDDDAPGQLTRAALERWG
MSMEG_5632|M.smegmatis_MC2_155 VGRDKERLAELAQTIDAP----DRVATLALDLTDDDAPQRIVDAAVTRWG
TH_4328|M.thermoresistible__bu TGRDSGRLAGLEKTLRKAGVPADRIATLAVDLTADEAPQRIVDAAVSRWG
Mflv_1783|M.gilvum_PYR-GCK TGRDAGRLAELEKAAPEP----GRIATLAVDLTDDDAPQRIVSAALDRWG
Mvan_4967|M.vanbaalenii_PYR-1 TGRDPGRLAALEKSS-------DRIATLAVDLTDDDAPQRIVSAALERWG
MAV_5213|M.avium_104 TGRDPKRLAELETAIGDP----DRVATVAVDLTDDDAPRRIVDRALQRWG
MAB_4632|M.abscessus_ATCC_1997 ASRTESQLREVAAKVQAAG---RQAEVVVADLSDTEASAALAQKAVDRFG
Mb2882c|M.bovis_AF2122/97 GDVDVEAGGAAADELSGLF--------VPTDVCDEDAVNGLFDGAAETYG
Rv2857c|M.tuberculosis_H37Rv GDVDVEAGGAAADELSGLF--------VPTDVCDEDAVNGLFDGAAETYG
MLBr_01807|M.leprae_Br4923 ----THRASVVPDWFFG----------VECDVTDNDAIGAAFKAVEEHQG
* :* . *
MUL_4820|M.ulcerans_Agy99 RIDFLINAAGEGSP--KSVHETDDESLDYVLSLMLRAPFRLTREVLPHLR
MMAR_0302|M.marinum_M RIDFLINAAGVGSP--KPVHETDDESLDYVLSLMLRAPFRLTREVLPHLR
MSMEG_5632|M.smegmatis_MC2_155 KVDFLINNAGVGSP--KPLHETDDESLDYFLGLMLRAPFRLARDVIPHMG
TH_4328|M.thermoresistible__bu HIDFLINNAGVGSP--KPLGETDDDTLDHFLGLMLRAPFRLARDVLPHMR
Mflv_1783|M.gilvum_PYR-GCK RIDFLVNNAGIGSP--KPLHETDDETLDHFLGVMLRAPFRLARDVIVHMQ
Mvan_4967|M.vanbaalenii_PYR-1 RIDFLVNNAGIGSP--KPLHETDDETLDRFLDIMLRAPFRLARDVIVHMQ
MAV_5213|M.avium_104 HIDFLINNAGVGSP--KPLHETDDETLDYFLNLMLRAPFRLAREVLPHLP
MAB_4632|M.abscessus_ATCC_1997 RLDVVVNNVGGTMP--KPFLDTTNDDLAEAFSFNVLSGHALLRAAVPHLL
Mb2882c|M.bovis_AF2122/97 RIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMV
Rv2857c|M.tuberculosis_H37Rv RIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMV
MLBr_01807|M.leprae_Br4923 PVEVLVANAGITS--DMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMI
::. . .* . .. : . . : . ::
MUL_4820|M.ulcerans_Agy99 PG--SAIINITSTFAVVG-GLRGGAYSAAKGGLAALTTHIACQYGAQGIR
MMAR_0302|M.marinum_M PG--SAIINITSTFAVVG-GLRGGAYSAAKGGLTALTTHIACQYGAQGIR
MSMEG_5632|M.smegmatis_MC2_155 PG--SAIINVTSTFAVVG-GLRGGAYSAAKGGLTALTTHIACQYGAQGIR
TH_4328|M.thermoresistible__bu PG--SAIINITSTFAVVG-GLRGGAYSAAKGGLTALTTHIACQYGAQGIR
Mflv_1783|M.gilvum_PYR-GCK PG--SAIINVTSTFAVVG-GLRGGAYSAAKGGLTALTSHIACQYGAQGIR
Mvan_4967|M.vanbaalenii_PYR-1 PG--SAIINITSTFAVVG-GLRGGAYSAAKGGLTALTTHIACQYGAQGIR
MAV_5213|M.avium_104 PG--SAIINVTSTFAVVG-GLRGGAYSAAKGGLTALTTHIACQYGASGIR
MAB_4632|M.abscessus_ATCC_1997 KSDNASVINITSTMGRLP-GRAFLGYCTAKGALAHYTRTAAMDLSPR-IR
Mb2882c|M.bovis_AF2122/97 LAGKGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIR
Rv2857c|M.tuberculosis_H37Rv LAGKGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIR
MLBr_01807|M.leprae_Br4923 KQRFGRYIFMGSVVGLSG-VPGQANYAASKSGLIGMARSLARELGSRSIT
. : * .. * ::*..: : . : . *
MUL_4820|M.ulcerans_Agy99 CNAVAPGVIQTAMTQHRLADERFRRLN-VEITPHQRLGTVDDVANTVAFL
MMAR_0302|M.marinum_M CNAVAPGVIQTAMTQHRLADERFRRIN-VEMTPHQRLGTVDDVANTVAFL
MSMEG_5632|M.smegmatis_MC2_155 CNAVAPGVTLTPMVATRLEDDRFRKIN-TEMTPHQRLGRVEDVASTVAFL
TH_4328|M.thermoresistible__bu CNAVAPGVTLTPMVATRLEDERFRKIN-TEMTPHQRLGRVEDIAATVAFL
Mflv_1783|M.gilvum_PYR-GCK CNAVAPGVTLTPMVATRLEDERFRKIN-TEMTPHQRLGRVEDIASTVAFL
Mvan_4967|M.vanbaalenii_PYR-1 CNAVAPGVTLTPMVASRLEDERFRKIN-TEMTPHQRLGRVEDVASTVAFL
MAV_5213|M.avium_104 CNAVAPGVTVTPMVENRLQDPRFRKIN-TEMTPHQRLGSVDDIAATVAFL
MAB_4632|M.abscessus_ATCC_1997 VNGIAPGSILTSALEVVAGNDEMRNTL-EQNTPLHRLGDPADIAAAAVYL
Mb2882c|M.bovis_AF2122/97 VNALCPGPVNTPLLQELFAKNPERAARRMVHVPLGRFAEPDEIAAAVAFL
Rv2857c|M.tuberculosis_H37Rv VNALCPGPVNTPLLQELFAKNPERAARRMVHVPLGRFAEPDEIAAAVAFL
MLBr_01807|M.leprae_Br4923 ANVVAPGFIDTEMTRAMDSKYQEAALQ---AIPLGRIGAVEEIAGAVSFL
* :.** * . * *:. ::* :. :*
MUL_4820|M.ulcerans_Agy99 CSEGGAFINGQTIVVDGG------------------------
MMAR_0302|M.marinum_M CSEGGAFINGQTIVVDGGWSSTKYLSDFALTAQWSPAQDDSK
MSMEG_5632|M.smegmatis_MC2_155 CSEGGSFINGQTIVVDGGWSSTKYLSDHALNSDWVPR-----
TH_4328|M.thermoresistible__bu CSPGAEFINGQTIVVDGGWSSTKYLSDHALTSEWVPR-----
Mflv_1783|M.gilvum_PYR-GCK CSDGGEFINGQTIVVDGGWSSTKYLSDFALNSEWVARQGTS-
Mvan_4967|M.vanbaalenii_PYR-1 CSEGGSFINGQTIVVDGGWSTTKYLSDFALNAHWVER-----
MAV_5213|M.avium_104 CSPGGSFINGQTIVVDGGWSSTKYLSEYALTSEWIAP-----
MAB_4632|M.abscessus_ATCC_1997 ASPAGSFLTGKVLEVDGGLIAPNLDIPLPDLA----------
Mb2882c|M.bovis_AF2122/97 ASDDASFITASTFLVDGGISSAYVTPL---------------
Rv2857c|M.tuberculosis_H37Rv ASDDASFITASTFLVDGGISSAYVTPL---------------
MLBr_01807|M.leprae_Br4923 ASEDAGYISGAVIPVDGGLGMGH-------------------
.* . ::.. .: ****