For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSLLGFGRGERRTRKVDAVVTGAASGIGRAFAVELARRGGRVVCADVDLPGAKETAALIGERAIEAACDV SVFDEVEALADTAEEWFGKPSDLVINNAGIGAGGQVVGAAPLADWQKTLGVNLWGVIHGSHVFAPRLRAL GRGGIINVASAASFAAAPRMGAYNVSKAGVLALSETLAAELSGTGVAVTVLCPTFVKTSIVDSGIIEESA ARLAATAMKWTGVSADSVARGCLDAHDRGQLYVLPQFDARFMWQAKRLAPDTYTKALGIAERFSRQRGN*
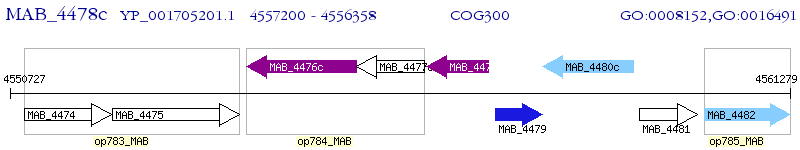
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4478c | - | - | 100% (280) | putative short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_2071 | - | 3e-97 | 64.00% (275) | short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_4053c | - | 1e-85 | 61.80% (267) | putative short chain dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0874c | - | 2e-35 | 39.18% (245) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4610 | - | 1e-104 | 68.35% (278) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0851c | - | 2e-35 | 39.18% (245) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 5e-33 | 43.15% (197) | short chain dehydrogenase |
| M. marinum M | MMAR_3141 | - | 6e-97 | 64.71% (272) | short-chain membrane-associated dehydrogenase |
| M. avium 104 | MAV_1797 | - | 3e-90 | 61.99% (271) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_4280 | - | 2e-32 | 37.55% (261) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4323 | - | 9e-33 | 43.62% (188) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3643 | - | 4e-87 | 59.34% (273) | short-chain membrane-associated dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1521 | - | 2e-85 | 60.37% (270) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4610|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3141|M.marinum_M --------------------------------------------------
MAB_4478c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_1797|M.avium_104 --------------------------------------------------
MUL_3643|M.ulcerans_Agy99 --------------------------------------------------
Mvan_1521|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
TH_4323|M.thermoresistible__bu --------MATEHFIDSSGGVRIAAYEEGNPDGPTVVLVHGWPDSHVVWD
MSMEG_4280|M.smegmatis_MC2_155 ----------MYTFVTSQDGTRIAVYQQGDPERPTLVMAHGWPDSHVLWD
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4610|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3141|M.marinum_M --------------------------------------------------
MAB_4478c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_1797|M.avium_104 --------------------------------------------------
MUL_3643|M.ulcerans_Agy99 --------------------------------------------------
Mvan_1521|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
TH_4323|M.thermoresistible__bu SVAPLLADRFRIIRYDNRGVGRTTAPRRVADHTMARYADDFGAVIDALAP
MSMEG_4280|M.smegmatis_MC2_155 GVVGELGDRYRIVRYDNRGAGASDVPKATSAYTMERFADDFSAVIDAVSP
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4610|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3141|M.marinum_M --------------------------------------------------
MAB_4478c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_1797|M.avium_104 --------------------------------------------------
MUL_3643|M.ulcerans_Agy99 --------------------------------------------------
Mvan_1521|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
TH_4323|M.thermoresistible__bu GQRVHVLAHDWGSAGVWEYLARPEAEDRIASFTSVSGPSGDHLARYVLDG
MSMEG_4280|M.smegmatis_MC2_155 DAPVHVLAHDWGSVGIWEYLSRQGSAARMVSFTSVSGPSTDHYGAYVRRG
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4610|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3141|M.marinum_M --------------------------------------------------
MAB_4478c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_1797|M.avium_104 --------------------------------------------------
MUL_3643|M.ulcerans_Agy99 --------------------------------------------------
Mvan_1521|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIR-RTM
TH_4323|M.thermoresistible__bu LAAPYRPRRFLRSLTQLLRLSYMGFFSIPVLAPLLVRGVLSKALRRSLRL
MSMEG_4280|M.smegmatis_MC2_155 LARPYRPVQFARAVNLASRFSYWIPFSIPVLAPALMR----RGAARRLQA
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4610|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3141|M.marinum_M --------------------------------------------------
MAB_4478c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_1797|M.avium_104 --------------------------------------------------
MUL_3643|M.ulcerans_Agy99 --------------------------------------------------
Mvan_1521|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 VDNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPV
TH_4323|M.thermoresistible__bu RDGVPADRLHHSDTFAADAARSIKVYGANFLRTLRHARTDH---FVRVPV
MSMEG_4280|M.smegmatis_MC2_155 ID--TPGPKFQSENLDIDAANSLKIYRANAIRSFTHVRRDH---YVQVPV
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_4610|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3141|M.marinum_M --------------------------------------------------
MAB_4478c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_1797|M.avium_104 --------------------------------------------------
MUL_3643|M.ulcerans_Agy99 --------------------------------------------------
Mvan_1521|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 QLIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVH
TH_4323|M.thermoresistible__bu QLIVNTRDPYVRPHVYDETDRWVSRLWRRDIKAGHWSPMSHPQVLATAVT
MSMEG_4280|M.smegmatis_MC2_155 QLICNRKDPVVRGEGYRDESKWVPLLWRRDLDAGHWAPFSHPRAIAQAVG
Mb0874c|M.bovis_AF2122/97 -----------------------MDGFPGRGAVITGGASGIGLATGTEFA
Rv0851c|M.tuberculosis_H37Rv -----------------------MDGFPGRGAVITGGASGIGLATGTEFA
Mflv_4610|M.gilvum_PYR-GCK --------------MDILRWDRPPRLSRRANAVVTGAGSGIGRAFAVELA
MMAR_3141|M.marinum_M --------------MRFLPFGSSPGRTHRAHAVVTGAGSGIGRAFAVELA
MAB_4478c|M.abscessus_ATCC_199 -------------MVSLLGFGRGERRTRKVDAVVTGAASGIGRAFAVELA
MAV_1797|M.avium_104 --------------MLDRLLHRS-KTSRGALAVVTGAGSGIGAAFALELG
MUL_3643|M.ulcerans_Agy99 -----------MLGPLDKLLGKSRKTSHDALAVVTGAGSGIGAAFALELG
Mvan_1521|M.vanbaalenii_PYR-1 ------------------MGLFGAKRTHRADAVITGAGSGIGAAFAVELA
MLBr_00862|M.leprae_Br4923 DLADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALA
TH_4323|M.thermoresistible__bu ELADHLAGAPAGRALLRAQVGRPREYFGDMLVAITGAGGGIGRETALAFA
MSMEG_4280|M.smegmatis_MC2_155 EFVDHMEGAPPARALRRAEVARPRKAFSDMLVSVTGAASGIGRATALEFA
. :**...*** . :.
Mb0874c|M.bovis_AF2122/97 RRGARVVLGDVDKPGLRQAVNHLRAEGFDVHGVMCDVRHREEVTHLADEA
Rv0851c|M.tuberculosis_H37Rv RRGARVVLGDVDKPGLRQAVNHLRAEGFDVHSVMCDVRHREEVTHLADEA
Mflv_4610|M.gilvum_PYR-GCK RRGGRVVCADIDPVRAKETVGLVVQAGGEGLDIACDVTDEEQVRELADSA
MMAR_3141|M.marinum_M RRGGRVVCADKDPITAKESAELVRQAGGEGFDVVCDVTDLEQVRNLADAS
MAB_4478c|M.abscessus_ATCC_199 RRGGRVVCADVDLPGAKETAALIGER---AIEAACDVSVFDEVEALADTA
MAV_1797|M.avium_104 KRGGTVVCSDIDQAAAQRTADAITEHGAKALATRCDVSQFGVVQALAEQS
MUL_3643|M.ulcerans_Agy99 RRGSAVVCSDIDEVAAQNTVAALTEAGARAISIGCDVSRIEDVTALAERS
Mvan_1521|M.vanbaalenii_PYR-1 RRGGRVVCSDVSVEAARGTADAIIADGGRALPAQCDVTDIDAMYRLAGEA
MLBr_00862|M.leprae_Br4923 RRGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQV
TH_4323|M.thermoresistible__bu REGAEVIVSDIDEATAKDTAAEIAARGGVAHAYQLDVSDAAAVDRFADEV
MSMEG_4280|M.smegmatis_MC2_155 RLGAEIVVSDIDEAGAKATATAIDEAGGVAHAYGLDVADPDAVQRFVDDV
: *. :: .* . : :. : ** : :.
Mb0874c|M.bovis_AF2122/97 FRLLG-HFDVVFSNAGIVVGG-PIVEMTHDDWRWVIDVDLWGSIHTVEAF
Rv0851c|M.tuberculosis_H37Rv FRLLG-HVDVVFSNAGIVVGG-PIVEMTHDDWRWVIDVDLWGSIHTVEAF
Mflv_4610|M.gilvum_PYR-GCK EKWFGAAASLVINNAGIGIGGNVIGATPKRDWEATLSVNLWGVIYGCEIF
MMAR_3141|M.marinum_M EDWFGKAASLVINNAGIGAGGNRIGATSVEDWNAAISVNLWGVIYGCETF
MAB_4478c|M.abscessus_ATCC_199 EEWFGKPSDLVINNAGIGAGGQVVGAAPLADWQKTLGVNLWGVIHGSHVF
MAV_1797|M.avium_104 QSWFGAPPTLVINNAGVGAGGAAIGDAPLDDWQWTLGINLWGPIHGCHVF
MUL_3643|M.ulcerans_Agy99 QAWFDGPPTLVINNAGVGAGGAGIGEIPLDDWLWTLGINLWGPIHGCHVF
Mvan_1521|M.vanbaalenii_PYR-1 QTWFDGPPTLVINNAGVGAGGAFIGETELADWKWVLDINLWGPIHGCHVF
MLBr_00862|M.leprae_Br4923 SAKHG-VPDIVVNNAGIGQAG-RFLDTPPEEFDRVLAVNLGGVVNCCRAF
TH_4323|M.thermoresistible__bu CARHG-VPDIVVNNAGIGHAG-RFLDTPRDQWDRVLEVNLGGVVNGCRAF
MSMEG_4280|M.smegmatis_MC2_155 CATHG-VPDIVVNNAGIGHAG-NFLDTPADQYERVLEVNFGGVVNCCRAF
. :*..***: .* . :: .: ::: * : . *
Mb0874c|M.bovis_AF2122/97 LPRLLEQGTG---GHVVFTASFAGLVPNAGLGAYGVAKYGVVGLAETLAR
Rv0851c|M.tuberculosis_H37Rv LPRLLEQGTG---GHVVFTASFAGLVPNAGLGAYGVAKYGVVGLAETLAR
Mflv_4610|M.gilvum_PYR-GCK VPRLRAKGS----GGIINVASAASFGAAPRMGAYNVSKAGVLSLSETLAA
MMAR_3141|M.marinum_M VPRLRSNGR----GGVINVASAASFGSAPRMGAYNVSKAGVLALSETLAA
MAB_4478c|M.abscessus_ATCC_199 APRLRALGR----GGIINVASAASFAAAPRMGAYNVSKAGVLALSETLAA
MAV_1797|M.avium_104 TPILRDAAPSAAPRGIINVASAAAFGAAPGMAAYNVSKAGVLSLSETLAA
MUL_3643|M.ulcerans_Agy99 TPILREAGRAVGPRGIINVASAAAFGAAPGMAAYNVSKAGVLSLSETLAA
Mvan_1521|M.vanbaalenii_PYR-1 APILREAGR---PAGIINVASAAAFGAAPGMAAYNVSKAGTLSLSETLAA
MLBr_00862|M.leprae_Br4923 GQRLVERGTG---GHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRA
TH_4323|M.thermoresistible__bu APRLVERGTG---GHIVNIASMASYAPLQSLNAYCTSKAAVYMFSDCLRA
MSMEG_4280|M.smegmatis_MC2_155 GRRLVEQGTG---GHIVNVASMAAYSPAGSMNAYATSKAAVFMFSDCLRA
* . :: :* *. . : ** .:* .. ::: *
Mb0874c|M.bovis_AF2122/97 EVTADGIGVSVLCPMVVETNLVANSERIRGAACAQSSTTGSPGPLPLQDD
Rv0851c|M.tuberculosis_H37Rv EVTADGIGVSVLCPMVVETNLVANSERIRGAACAQSSTTGSPGPLPLQDD
Mflv_4610|M.gilvum_PYR-GCK ELSGTGVAVTVLCPTFVKTNIVDNPGIEESAAKLATNLMKWTG-------
MMAR_3141|M.marinum_M ELSGTNVNVTVLCPTFVKTNIAKNPQIEESAAKLATNLMRWTG-------
MAB_4478c|M.abscessus_ATCC_199 ELSGTGVAVTVLCPTFVKTSIVDSGIIEESAARLAATAMKWTG-------
MAV_1797|M.avium_104 ELSGTPVRVTVLCPTFVKTTILESGRISEESGELAAKLMRWTG-------
MUL_3643|M.ulcerans_Agy99 ELSGTGINVTVLCPTFVKTNIVESGRISEQSSEVVTKLIRWTG-------
Mvan_1521|M.vanbaalenii_PYR-1 ELAGTGINVTVLCPTFVKTNIVESGRISTGTSQLAERLMRWTG-------
MLBr_00862|M.leprae_Br4923 ELDAVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRL
TH_4323|M.thermoresistible__bu ELDSAGIMLTTICPSTVNTNIIGSTRFNAPDGLDDVVDTRREQLQKLFAL
MSMEG_4280|M.smegmatis_MC2_155 ELAHAGIGVTTLCPGMIDTGIVDTTRFDVPAHKAGQVEARRAQVKKTFQA
*: : ::.:** :.* : .
Mb0874c|M.bovis_AF2122/97 N-LGVDDIAQLTADAILANRLYVLPHAASRASIRRRFERIDRTFDEQAAE
Rv0851c|M.tuberculosis_H37Rv N-LGVDDIAQLTADAILANRLYVLPHAASRASIRRRFERIDRTFDEQAAE
Mflv_4610|M.gilvum_PYR-GCK --VSAESVARKTLDANDRGQLHVLPQVD--AKILWLFKRAVPATYTRALG
MMAR_3141|M.marinum_M --ISPDQVARTTLNAHDRGQIYVVPQLD--AKILWQLKRALPGQFTRALG
MAB_4478c|M.abscessus_ATCC_199 --VSADSVARGCLDAHDRGQLYVLPQFD--ARFMWQAKRLAPDTYTKALG
MAV_1797|M.avium_104 --FSADKVARICLDAHDRGDLYCMPQLD--AQIGWHIKRLAPQAYTRAAG
MUL_3643|M.ulcerans_Agy99 --FSPDKVARICLDANDRGDLYCMPQLD--AKIGWNIKRMAPGTYTRAAG
Mvan_1521|M.vanbaalenii_PYR-1 --FPAARVARSALDTLDRGGLYCMPQPE--ARIGWGVKRFTPTVFTRAAG
MLBr_00862|M.leprae_Br4923 RRYGPDKVADTILSAIKKNKPIRPVAPE--AYALYGISRVLPQVLRSTAR
TH_4323|M.thermoresistible__bu RRFGPDKAAKAIVSAVKQNKPIRPVMPE--AYFVYGVSRVLPQALRSTAR
MSMEG_4280|M.smegmatis_MC2_155 RRYGPEKVAKAVTSAVSRNQALRPVTPE--AYVAYGVSRLFPAALRRAAR
* .: . * .* :
Mb0874c|M.bovis_AF2122/97 GWRH-------------
Rv0851c|M.tuberculosis_H37Rv GWRH-------------
Mflv_4610|M.gilvum_PYR-GCK LVERIAR----------
MMAR_3141|M.marinum_M LVERVASWNDPAEQREQ
MAB_4478c|M.abscessus_ATCC_199 IAERFSRQRGN------
MAV_1797|M.avium_104 LVSRINLP---------
MUL_3643|M.ulcerans_Agy99 LILRATMR---------
Mvan_1521|M.vanbaalenii_PYR-1 IATRITT----------
MLBr_00862|M.leprae_Br4923 IRVI-------------
TH_4323|M.thermoresistible__bu GRVL-------------
MSMEG_4280|M.smegmatis_MC2_155 SQVL-------------