For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTQTVLITGANGGLGRLMRPRLAREGRTLRLLDLVTPEPPADGEDVEVLQGSVTDEKVVRDACDGVDAVI HLGGISVEAPWQDILTNNIDGTRVLLEQARDAGVERVVLASSNHAVGFYGKEEAGPDGLRADILPRPDTY YGFSKAALEALGSLFHSRYGMDVTCLRIGSCFEKPWDARSLWTWMSPDDGARLLEACLATPEPGFRIVWG ISRNTRRWWSLEAGEQIGYHPQDDSEVFAQEILAHVEESGQGADVRLVGGKFCFTELGAHQH*
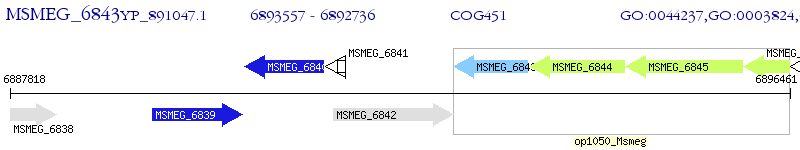
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6843 | - | - | 100% (273) | NAD-dependent epimerase/dehydratase |
| M. smegmatis MC2 155 | MSMEG_5228 | - | 2e-09 | 35.00% (120) | 3-beta hydroxysteroid dehydrogenase/isomerase family protein |
| M. smegmatis MC2 155 | MSMEG_5976 | - | 7e-08 | 34.39% (157) | epimerase/dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1136c | - | 2e-11 | 35.42% (144) | cholesterol dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2075 | - | 9e-11 | 35.11% (131) | 3-beta hydroxysteroid dehydrogenase/isomerase |
| M. tuberculosis H37Rv | Rv1106c | - | 2e-11 | 35.42% (144) | cholesterol dehydrogenase |
| M. leprae Br4923 | MLBr_01942 | - | 2e-11 | 35.83% (120) | putative cholesterol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1344c | - | 2e-11 | 29.21% (178) | putative dTDP-glucose-4,6-dehydratase-related protein |
| M. marinum M | MMAR_3324 | - | 1e-11 | 38.66% (119) | nucleoside-diphosphate-sugar epimerase |
| M. avium 104 | MAV_1225 | - | 8e-11 | 33.33% (144) | 3-beta hydroxysteroid dehydrogenase/isomerase family protein |
| M. thermoresistible (build 8) | TH_3827 | - | 1e-11 | 33.52% (182) | PUTATIVE putative dehydrogenase |
| M. ulcerans Agy99 | MUL_0172 | - | 4e-11 | 35.83% (120) | cholesterol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4635 | - | 4e-10 | 34.81% (135) | 3-beta hydroxysteroid dehydrogenase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1136c|M.bovis_AF2122/97 MLRRMGDA-SLTTELGRVLVTGGAGFVGANLVTTLLDRGHWVRSFDRAPS
Rv1106c|M.tuberculosis_H37Rv MLRRMGDA-SLTTELGRVLVTGGAGFVGANLVTTLLDRGHWVRSFDRAPS
MAV_1225|M.avium_104 MIGPMGDP-SLTAELGRVLVTGGSGFVGANLVTTLLERGYQVRSFDRAPS
MUL_0172|M.ulcerans_Agy99 MLRPMGDA-SLTTELGRVLVTGGSGFVGSNLVTTLLDRGHHVRSFDRAPS
MLBr_01942|M.leprae_Br4923 MLRGVGNA-SLTTELGRVLVTGGSGFVGANLVTALLERGYQVRSFDRAPM
Mflv_2075|M.gilvum_PYR-GCK ---------MLRTELGRVLVTGGSGFVGANLVTELLDRGLHVRSFDRVAS
Mvan_4635|M.vanbaalenii_PYR-1 ----MGDATTPTTELGRVLVTGGSGFVGANLVTELLDRGLQVRSFDRVPS
MMAR_3324|M.marinum_M MSTDQSARPAPFFAQRKIFITGANGFIGANLAARLRQLGARVTGVD----
TH_3827|M.thermoresistible__bu -MTDEDRIPCGAMSRPSVFITGANGFIGRSLGAHFAENGYDVRGVD----
MSMEG_6843|M.smegmatis_MC2_155 ------------MMTQTVLITGANGGLGRLMRPRLAREGRTLRLLDLVTP
MAB_1344c|M.abscessus_ATCC_199 ------------MSERRFLVTGSSGHLGEALVRTLRRDGVDVVGID----
.::**. * :* : : * : .*
Mb1136c|M.bovis_AF2122/97 LLPAHP-QLEVLQGDITDADVCAAAVDG----IDTIFHTAAIIELMGGAS
Rv1106c|M.tuberculosis_H37Rv LLPAHP-QLEVLQGDITDADVCAAAVDG----IDTIFHTAAIIELMGGAS
MAV_1225|M.avium_104 PLPAHP-HLEVLQGDITDAGVCAAAVEG----IDTIFHTAAIIDLMGGAS
MUL_0172|M.ulcerans_Agy99 PLPPHP-QLEVLQGDITDAATCATAVDG----VDTVFHTAAIIELMGGAS
MLBr_01942|M.leprae_Br4923 PLPQHP-QLEVLQGDITDATVCTTAMDS----IDTVFHTAAIIELMGGAS
Mflv_2075|M.gilvum_PYR-GCK ALPAHA-RLEIVEGDITDADDVAAAVEG----VDTVFHTAAIIDLMGGAS
Mvan_4635|M.vanbaalenii_PYR-1 PLPDHP-GLEVVQGDITDVDDVARAVGTGADKADTVFHTAAIIDLMGGAS
MMAR_3324|M.marinum_M -LVADP-ANGIIAGSTADPAAWASALDG----VDAVVHLAALVSTVVAV-
TH_3827|M.thermoresistible__bu -LAADP-ARNVVAGDVVNPRPWKSHLQG----CAVVLHTAAVVSNAVAA-
MSMEG_6843|M.smegmatis_MC2_155 EPPADGEDVEVLQGSVTDEKVVRDACDG----VDAVIHLGGISVEAPWQ-
MAB_1344c|M.abscessus_ATCC_199 -IAAGP--ATDIVGSIADRGLVAEAMSG----MTSVLHTATLHKPHIGSH
: *. .: :.* . :
Mb1136c|M.bovis_AF2122/97 VTDEYRQRSFAVNVGGTENLLHAGQRAGVQRFVYTSSNSVVMGGQNIAGG
Rv1106c|M.tuberculosis_H37Rv VTDEYRQRSFAVNVGGTENLLHAGQRAGVQRFVYTSSNSVVMGGQNIAGG
MAV_1225|M.avium_104 VTEEYRQRSFAVNVGGTENLVRAGQAAGVQRFVYTSSNSVVMGGQNIVGG
MUL_0172|M.ulcerans_Agy99 VTDEYRKRSFSVNVGGTENLVRAGQQAGVKRFVYTSSNSVVMGGQIIAGG
MLBr_01942|M.leprae_Br4923 VTDEYRQRSYTVNVGGTENLLRAGQKSGVKRFVYTASNSVVMGGTPITGG
Mflv_2075|M.gilvum_PYR-GCK VSEEYRQRSFAVNVTGTQNLVHAAQKAGVTRFVYTASNSVVMGGQRIAGG
Mvan_4635|M.vanbaalenii_PYR-1 VTEEYRQRSFAVNVTGTKNLVHAAQKAGVQRFVYTASNSVVMGGQRIAGG
MMAR_3324|M.marinum_M ------ETAWDVNVLGTKKVIDAAVDAGVRRFVHLSSIAAYG-WDFPDHV
TH_3827|M.thermoresistible__bu ------GQQWQANVLGTRRVLEAAARAGVERVVHFSSVRAYGDLGFPDGV
MSMEG_6843|M.smegmatis_MC2_155 -------DILTNNIDGTRVLLEQARDAGVERVVLASSNHAVGFYGKEEAG
MAB_1344c|M.abscessus_ATCC_199 P----RTDFVETNIEGTLSVLEAAVAAGVRSFVYTSTTSAFGHALVGARE
*: ** :: . :** .* :: .
Mb1136c|M.bovis_AF2122/97 DETLPYTDRFN--DLYTETKVVAERFVLAQNGV---DGMLTCAIRPSGIW
Rv1106c|M.tuberculosis_H37Rv DETLPYTDRFN--DLYTETKVVAERFVLAQNGV---DGMLTCAIRPSGIW
MAV_1225|M.avium_104 DETLPYTDRFN--DLYTETKVLAERFVLGQNGV---DGMLTCAIRPSGIW
MUL_0172|M.ulcerans_Agy99 DETLPYTNRFN--DLYTETKVVAEKFVLSQNGN---NEMLTCAIRPSGIW
MLBr_01942|M.leprae_Br4923 DETMPYTKRFN--DLYTETKVVAEKFVLSQNGVPDGETMLTCSIRPSGIW
Mflv_2075|M.gilvum_PYR-GCK DETLPYTERFN--DLYTETKVVAEKFVLSQNGV---SGLLTCSIRPSGIW
Mvan_4635|M.vanbaalenii_PYR-1 DETLPYTERFN--DLYTETKVVAEKFVLSQNGV---SGMLTCSIRPSGIW
MMAR_3324|M.marinum_M TEDYPTRVTGGL-STYVDTKTNSELVALAN-AN---RGMEMVVVRPADVY
TH_3827|M.thermoresistible__bu KETWPLRADG---RAYVDTKVAAEATAFAAHAS---GEIAVTVVRPGDVY
MSMEG_6843|M.smegmatis_MC2_155 PDGLRADILPRP-DTYYGFSKAALEALGSLFHSR--YGMDVTCLRIGSCF
MAB_1344c|M.abscessus_ATCC_199 AAWITEAVASVPKNIYGATKTAAEDIAHVVHQD---SGLPVIVLRTSRFF
* . : : :* . :
Mb1136c|M.bovis_AF2122/97 GNGDQTMFRKLFESVLKGHVKVLVGRKSARLDNSYVHNLIHGFILAAAHL
Rv1106c|M.tuberculosis_H37Rv GNGDQTMFRKLFESVLKGHVKVLVGRKSARLDNSYVHNLIHGFILAAAHL
MAV_1225|M.avium_104 GRGDQTMFRKLFESVIAGHVKVLIGRKSARLDNSYVHNLIHGFILAAQHL
MUL_0172|M.ulcerans_Agy99 GTGDQLMFRKLFESVIKGHVKVLVGPKSVLLDNSYVDNLIHGFMLAAQHL
MLBr_01942|M.leprae_Br4923 GRGDQTMFRKAFESVVSGHVKVLIGSKNAKLDNSYVHNLVHGLILAAEHL
Mflv_2075|M.gilvum_PYR-GCK GRGDQTMFRKVFESVLAGHVKVLVGNKETKLDNSYVHNLVHGFILAAEHL
Mvan_4635|M.vanbaalenii_PYR-1 GRGDQTMFRKVFESVLAGHVKVLVGNKNVKLDNSYVHNLVHGFILAAQHL
MMAR_3324|M.marinum_M GPGS-VWIREPIAMAKANQLILPERG-SGVFDVIYIDNFVDAMVLVLATE
TH_3827|M.thermoresistible__bu GPGSRPWTIIPVEEIKKGRLILPAGG-QGVFSPVYIDDLIAGILLAATHP
MSMEG_6843|M.smegmatis_MC2_155 EKPWDARSLWTWMSPDDGARLLEACLATPEPG----FRIVWGISRNTRRW
MAB_1344c|M.abscessus_ATCC_199 PEQD------DNDRVRNRYRDGNVKANEYLYRRVDLADVVDAHLLAAERG
.: .
Mb1136c|M.bovis_AF2122/97 VPDGTAPGQAYFINDAEPINMFEFARPVLEACGQRWPKMRISGPAVRWVM
Rv1106c|M.tuberculosis_H37Rv VPDGTAPGQAYFINDAEPINMFEFARPVLEACGQRWPKMRISGPAVRWVM
MAV_1225|M.avium_104 TPGGTAPGQAYFINDAEPINMFEFARPVVEACGVNWPRVRVNGPIVRAAM
MUL_0172|M.ulcerans_Agy99 VPGGSAPGQAYFINDAEPINMFDFARPVVEACGEKWPRVRVSGPMVHRAM
MLBr_01942|M.leprae_Br4923 VPGGTAPGQAYFINDGEPINFFDFMGPIIKACGENWPRVRISGRLVRNVM
Mflv_2075|M.gilvum_PYR-GCK VEGGSAPGQAYFINDGEPINMFEFARPVVQACGEPFPKFRVPGRLVWFAM
Mvan_4635|M.vanbaalenii_PYR-1 VDGGTAPGQAYFINDGEPINMFEFARPVMEACGEPWPTFRVSGRLVWFAM
MMAR_3324|M.marinum_M G----IAGEVFNLGEELAVSCQEYFGEVASWTG-AKVRS-VPIRIGAPAL
TH_3827|M.thermoresistible__bu D----AAGQDFNLPGVEPVTCKTFFGHYTRMLNKPPPRT-VPTRVAIALA
MSMEG_6843|M.smegmatis_MC2_155 WSLEAGEQIGYHPQDDSEVFAQEILAHVEESGQGADVRL-VGGKFCFTEL
MAB_1344c|M.abscessus_ATCC_199 PEIGWAKYVISATTPFSPGDAAQLRTDPGAVVARYFPEQ--PGLYAARGW
Mb1136c|M.bovis_AF2122/97 TGWQRLHFRFGFPAPLLEPLAVERLYLDNYFSIAKARRDLGYEPLFTTQQ
Rv1106c|M.tuberculosis_H37Rv TGWQRLHFRFGFPAPLLEPLAVERLYLDNYFSIAKARRDLGYEPLFTTQQ
MAV_1225|M.avium_104 TGWQRLHFRFGIPAPLLEPLAVERLYLDNFFSIAKASRDLGYQPLFTTEQ
MUL_0172|M.ulcerans_Agy99 TGWQRLHFRFGLPAPLLEPLAVERLYLDNYFSVDKARRDLGYEPKFTTEQ
MLBr_01942|M.leprae_Br4923 AVWQRLHFGFGLPKPPMEPLAVERVYLDNYFSIEKAHKELGYRPLFTTEQ
Mflv_2075|M.gilvum_PYR-GCK TIWQFLHFKFGLPKPLLEPLAVERLYLDNYFSIAKAQRDLGYQPLFTTEQ
Mvan_4635|M.vanbaalenii_PYR-1 TIWQFLHFRFGLPKPLLEPLAVERLYLDNYFSIAKAERDLGYRPLYTTEQ
MMAR_3324|M.marinum_M GAIGRIQRRLGMSSE-LGPALLHMLNRRYVVSNDKARDRLGFKPVVSYHE
TH_3827|M.thermoresistible__bu TAAGTVERFRGRSSE-LNAETVYYLTRAGGYSRAKAERVLGWVPQVSLDE
MSMEG_6843|M.smegmatis_MC2_155 GAHQH---------------------------------------------
MAB_1344c|M.abscessus_ATCC_199 SMFPTIDR---------------------VYVNARARDDLGWQPRYDYRR
Mb1136c|M.bovis_AF2122/97 ALTECLPYYVSLFEQMKNEARAEKTAATVKP----
Rv1106c|M.tuberculosis_H37Rv ALTECLPYYVSLFEQMKNEARAEKTAATVKP----
MAV_1225|M.avium_104 AMSECLPYYVGMFEQMKRQALAGKASA--------
MUL_0172|M.ulcerans_Agy99 ALKECLPYYVDLFRQMKSEVKAGGR----------
MLBr_01942|M.leprae_Br4923 AMAECLPYYTELFEQIKVAAKPHLASIAATPRPE-
Mflv_2075|M.gilvum_PYR-GCK ALAQCIPYYVELFDRMKREAGNPVVQAAAPAPPKG
Mvan_4635|M.vanbaalenii_PYR-1 ALEHCIPYYVDLFGRMKAEAAQPAVQAAAPVPPAG
MMAR_3324|M.marinum_M GMARSK-------EWARHEGLI-------------
TH_3827|M.thermoresistible__bu GMARTE-------AWLRTEGYLD------------
MSMEG_6843|M.smegmatis_MC2_155 -----------------------------------
MAB_1344c|M.abscessus_ATCC_199 ILDCLAAEQDFRSPLAREIGAKGYHDEPTGVYTT-