For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAEATAHEHTGDTADSKLARKITGPLLFLFILGDVLGAGVYALMGVLAGKVGGALWAPLLAALMLALLT AGSYAELVTKYPKAGGAAVFAERAFKQPLVSFLVGFSMLAAGVTSAAGLSLAFAGDYLATFIDVPAVPAA IVFLALVGALNARGISESVKSNVVMTVIELSGLLIVIIAGAVMVGGGRGDLGRLGEFPESTAPAVAILAG AIIAYYSFVGFETSANVAEEIRNPSKVYPAALFGSLITAGVVYVLVGMASAAVLAPDELAASSGPLLEVV SESGLGVPDWVFSAIALIAVANGALLTMIMASRLAFGMAEHRLLPSVLSRVLTKRRTPWVAIVATTAVAM VLTAIGELSTLAETVVLLLLFVFTATNIAVLVLRRDTVEHDHFRVWTAVPVLGVASCVLLMTQQTAKVWM FAGILMAVGVVLYFVARAATNRTADETV
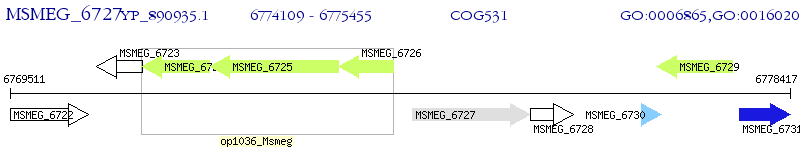
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6727 | - | - | 100% (448) | amino acid permease-associated region |
| M. smegmatis MC2 155 | MSMEG_5848 | - | 3e-87 | 42.66% (443) | amino acid permease-associated region |
| M. smegmatis MC2 155 | MSMEG_1838 | - | 3e-37 | 28.21% (475) | cationic amino acid transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3281c | - | 9e-33 | 26.96% (460) | cationic amino acid transport integral membrane protein |
| M. gilvum PYR-GCK | Mflv_4427 | - | 1e-179 | 72.65% (446) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv3253c | - | 9e-33 | 26.96% (460) | cationic amino acid transport integral membrane protein |
| M. leprae Br4923 | MLBr_01974 | - | 3e-13 | 24.56% (399) | putative permease |
| M. abscessus ATCC 19977 | MAB_2598 | - | 7e-35 | 28.95% (411) | hypothetical protein MAB_2598 |
| M. marinum M | MMAR_1290 | - | 9e-37 | 27.13% (457) | cationic amino acid transport integral membrane protein |
| M. avium 104 | MAV_4216 | - | 2e-34 | 26.48% (457) | cationic amino acid transporter |
| M. thermoresistible (build 8) | TH_4382 | - | 5e-87 | 42.89% (443) | PUTATIVE amino acid permease-associated region |
| M. ulcerans Agy99 | MUL_2597 | - | 1e-36 | 27.13% (457) | cationic amino acid transport integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_5147 | - | 1e-88 | 42.56% (437) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6727|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4427|M.gilvum_PYR-GCK --------------------------------------------------
TH_4382|M.thermoresistible__bu --------------------------------------------------
Mvan_5147|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3281c|M.bovis_AF2122/97 --------------------------------------------------
Rv3253c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1290|M.marinum_M --------------------------------------------------
MUL_2597|M.ulcerans_Agy99 --------------------------------------------------
MAV_4216|M.avium_104 --------------------------------------------------
MAB_2598|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01974|M.leprae_Br4923 MFLSADVSCIEIVDINCSELRNDRRIPPQLALFCTNAANKAESAVTSHGQ
MSMEG_6727|M.smegmatis_MC2_155 ---------------------------------------MTAEATAHEHT
Mflv_4427|M.gilvum_PYR-GCK -------------------------------------------MSVDTAE
TH_4382|M.thermoresistible__bu ---------------------------------------------MSGTR
Mvan_5147|M.vanbaalenii_PYR-1 -----------------------------------------MSESVAGPR
Mb3281c|M.bovis_AF2122/97 ----------------------------------MAGRRRMKSVEQSIAD
Rv3253c|M.tuberculosis_H37Rv ----------------------------------MAGRRRMKSVEQSIAD
MMAR_1290|M.marinum_M ----------------------------------MAGRLRTKSVEQSIAD
MUL_2597|M.ulcerans_Agy99 ----------------------------------MAGGLRTKSVEQSIAD
MAV_4216|M.avium_104 ----------------------------------MASRWRTKSVEQSIAD
MAB_2598|M.abscessus_ATCC_1997 -------------------------------------------------M
MLBr_01974|M.leprae_Br4923 LPIMANRRWAPHTRRQSGRATSTCSSLMRFSASESRAHSRFECYTLVRWS
MSMEG_6727|M.smegmatis_MC2_155 GDTADSKLARKITGPLLFLFILGDVLGAGVYALMGVLAGKVGG-ALWAPL
Mflv_4427|M.gilvum_PYR-GCK GTAEPTRLARRITGPLLFLFILGDVLGAGIYALMGELSSEVGG-VLWAPM
TH_4382|M.thermoresistible__bu TEQAQPELRRVLGPGLLLLFIVGDILGTGIYALVGDVAGEVGG-AAWLPF
Mvan_5147|M.vanbaalenii_PYR-1 SPEKQPELKRVMGPGLLLLFVVGDILGTGVYALVGDVAGEVGG-AAWLPF
Mb3281c|M.bovis_AF2122/97 TDEPTTRLRKDLTWWDLVVFGVSVVIGAGIFTVTASTAGDITGPAIWISF
Rv3253c|M.tuberculosis_H37Rv TDEPTTRLRKDLTWWDLVVFGVSVVIGAGIFTVTASTAGDITGPAIWISF
MMAR_1290|M.marinum_M TDEPETRLRKDLTWRDLVVFGVSVVIGAGIFTVTASTAGDITGPAIWISF
MUL_2597|M.ulcerans_Agy99 TDEPETRLRKDLTWRDLVVFGVSVVIGAGIFTVTASTAGDITGPAIWISF
MAV_4216|M.avium_104 TDEPDTRLRKDLTWWDLVVFGVAVVIGAGIFTVTASTTGDITGPAIWVSF
MAB_2598|M.abscessus_ATCC_1997 SSQQPGALRRELGTFDAVMIGLGSMIGAGIFAALAPAARSAGS-ALLIGL
MLBr_01974|M.leprae_Br4923 IRGQGGRAISKLGFWSVVMLGINSIIGAGIFMTPGEVIKIAGP-FAPIAY
: .:: : ::*:*:: .
MSMEG_6727|M.smegmatis_MC2_155 LAALMLALLTAGSYAELVTKYPKAGGAAVFAERAFKQPLVSFLVGFSMLA
Mflv_4427|M.gilvum_PYR-GCK TVALGLALLTAGSYAELVTKYPRAGGAAVFAERAFGKPLISFLVGFSMLA
TH_4382|M.thermoresistible__bu LIAFLIATVTAFSYLELVTKYPQAAGAALYVHKAFGIQFVTFLVAFIVMC
Mvan_5147|M.vanbaalenii_PYR-1 LVAFAIATVTAFSYLELVTKYPQAAGAALYAHKAFGIHFITFLVAFVVMC
Mb3281c|M.bovis_AF2122/97 LIAAATCALAALCYAEFASTLPVAGSAYTFSYATFG-EFLAWVIGWNLVL
Rv3253c|M.tuberculosis_H37Rv LIAAATCALAALCYAEFASTLPVAGSAYTFSYATFG-EFLAWVIGWNLVL
MMAR_1290|M.marinum_M LIAAGTCALAALCYAEFASTLPVAGSAYTFSYATFG-EFLAWVIGWNLVL
MUL_2597|M.ulcerans_Agy99 LIAAGTCALAALCYAEFASTLPVAGSAYTFSYATFG-EFLAWVIGWNLVL
MAV_4216|M.avium_104 VIAAGTCALAALCYAEFASTLPVAGSAYTFSYATFG-EFLAWIIGWNLLL
MAB_2598|M.abscessus_ATCC_1997 CVAAVIAYCNATSSARLAAVYPASGGTYVYGRERLG-DFWGYLAGWGFVV
MLBr_01974|M.leprae_Br4923 TLAGIFAAVLAIVFATAAKYVKTNGSSYAYTTAAFG-HRIGIYVGVTHAI
* . * . ..: : : .
MSMEG_6727|M.smegmatis_MC2_155 AGVTSAAGLSLAFAGDYLA-----------TFIDVPAVPAAIVFLALVGA
Mflv_4427|M.gilvum_PYR-GCK AGVTSAAGLALAFSGDYLA-----------TFVDVPVALAAVVFLALVAC
TH_4382|M.thermoresistible__bu SGITSASTASRFFAANLEVGLGQ-------EWGRLGVAGVALLFMALLAM
Mvan_5147|M.vanbaalenii_PYR-1 SGITSAATASRAFAANFFEGAGI-------DASKLGVVVLALLFMAALAA
Mb3281c|M.bovis_AF2122/97 ELAMGAAVVAKGWSSYLGTVFGFGNGTG--HLGSLQLDWGALVIVTLVAT
Rv3253c|M.tuberculosis_H37Rv ELAMGAAVVAKGWSSYLGTVFGFGNGTG--HLGSLQLDWGALVIVTLVAT
MMAR_1290|M.marinum_M ELAIGAAVVAKGWSSYLGTVFGFSGGTA--DFGSIRLDWGALLIVSIVSL
MUL_2597|M.ulcerans_Agy99 ELAIGAAVVAKGWSSYLGTVFGFSGGTA--DFGSIRLDWGALLSVSIVSL
MAV_4216|M.avium_104 ELAIGAAVVAKGWSSYLGTVFGFSGGTV--KFGAAQLDWGALVIVGGVAT
MAB_2598|M.abscessus_ATCC_1997 GKTASCAAMALTIGFYAWPG---------------HAHAVAVASVVALTA
MLBr_01974|M.leprae_Br4923 TASIAWGVLASSFVSTLLRVAFPEKIWADNDELISVKTLTFLVFIAVLLA
. . : : : :
MSMEG_6727|M.smegmatis_MC2_155 LNARGISESVKSNVVMTVIELSGLLIVIIAGAVMVGGG--RGDLGRLGEF
Mflv_4427|M.gilvum_PYR-GCK LNARGISESLKTNVVMTAIELSGLVLVIAVVAMMVGRG--DGEVGRVTEF
TH_4382|M.thermoresistible__bu VNLRGVGESVRLNVLLTIVEISGLLLVIFVGLWAFTGGGADVDFTRVVAF
Mvan_5147|M.vanbaalenii_PYR-1 VNYRGVGESVRLNVVLTIVEITGLVVVVLVGLWAFTGG-ADVDFSRITAF
Mb3281c|M.bovis_AF2122/97 LIALGTKLSSRFSAVVTAIKVSVVVLVVVVGAFYIRAANYSPFIPEPEVQ
Rv3253c|M.tuberculosis_H37Rv LIALGTKLSSRFSAVVTAIKVSVVVLVVVVGAFYIRAANYSPFIPEPEVQ
MMAR_1290|M.marinum_M LVAYGTKLSSRFSAVVTTIKVAVVILVVVVGAFYVKAANYSPFIPEPEAG
MUL_2597|M.ulcerans_Agy99 LVAYGTKLSSRFSAVVTTIKVAVVILVVVVGAFYVKAANYSPFIPEPEAG
MAV_4216|M.avium_104 LIALGTKLSSRFSAVITGIKVSVVVFVVVVGVFYIKRANYSPFIPKPEAG
MAB_2598|M.abscessus_ATCC_1997 INYVGVQKSAWLTRVIVAIVLSVLAAIVATAAGSGSVDIAALDLTGTS--
MLBr_01974|M.leprae_Br4923 INIFGNLVIRWANGISALGKIFALSVFIAGGLWTIITQHVNNYVTSRSAY
: * . : . : : .: .
MSMEG_6727|M.smegmatis_MC2_155 PES---------------TAP--AVAILAGAIIAYYSFVGFETSANVAEE
Mflv_4427|M.gilvum_PYR-GCK PDG---------------TTP--ALAILGGAILAYYSFVGFETSANVVEE
TH_4382|M.thermoresistible__bu ETT---------------TDKSVFLAVSAATSLAFFAMVGFEDSVNMAEE
Mvan_5147|M.vanbaalenii_PYR-1 ETG---------------EDKNTFLAVTAATSLAFFAMVGFEDSVNMAEE
Mb3281c|M.bovis_AF2122/97 HHGGGLDQSVFSLLTGAQGSHYGWYGVLAGASIVFFAFIGFDIVATMAEE
Rv3253c|M.tuberculosis_H37Rv HHGGGLDQSVFSLLTGAQGSHYGWYGVLAGASIVFFAFIGFDIVATMAEE
MMAR_1290|M.marinum_M PEGRGADQSLLSLLTGAPGSHYGWYGVLAGASIVFFAFIGFDIVATMAEE
MUL_2597|M.ulcerans_Agy99 PEGRGADQSLLSLLTGAPGSHYGWYGVLAGASIVFFAFIGFDIVATMAEE
MAV_4216|M.avium_104 GQAKGIDQSVLSLLTGAHTSHYGWYGVLAGASIVFFAFIGFDIVATMAEE
MAB_2598|M.abscessus_ATCC_1997 -----------------------WRGVLQAAGLLFFAFAGYARIATLGEE
MLBr_01974|M.leprae_Br4923 QPTTYSLLG------VTEIGKSTAASMTLATIVALYAFTGFESIANAAEE
.: .: : ::: *: .. **
MSMEG_6727|M.smegmatis_MC2_155 IRNPSKVYPAALFGSLITAGVVYVLVGMASAAVLAPDELAASSG---PLL
Mflv_4427|M.gilvum_PYR-GCK IRDPRRVYPRALFGALITAGVVYALVGLASAIAVPTDELSQSSG---PLL
TH_4382|M.thermoresistible__bu TRDPVVIFPRALLTGLGIAGAVYVVVAIVAVALVPVGTLQASDV---PLV
Mvan_5147|M.vanbaalenii_PYR-1 TKDPVNTFPKILLSGLSIAGVVYVLISIVAVALVPVGRLEESDT---PLV
Mb3281c|M.bovis_AF2122/97 TKRPQRDVPRGILASLGVVTLLYVAVSVVLSGMVPYTQLRTVPGRGPANL
Rv3253c|M.tuberculosis_H37Rv TKRPQRDVPRGILASLGVVTLLYVAVSVVLSGMVPYTQLRTVPGRGPANL
MMAR_1290|M.marinum_M TKHPQRDVPRGILVSLGVVTLLYVAVSIVLSGMVSYTQLRTVSGGGHANL
MUL_2597|M.ulcerans_Agy99 TKHPQRDVPRGILVSLGVVTLLYVAVSIVLSGMVSYTQLRTVSGGGHANL
MAV_4216|M.avium_104 TKHPQRDVPRGILASLGIVTLLYVAVAVVLSGMVSYTQLKTMPGRGQANL
MAB_2598|M.abscessus_ATCC_1997 VRDPARTIPRAIPIALGIALVVYVVVALAVLITLGPSRLASSAA---PLL
MLBr_01974|M.leprae_Br4923 MNTPDRTLPKAIPLTILTVGVIYILALVVAMLLGSDKIVATNHT------
. * * : : . :* :. :
MSMEG_6727|M.smegmatis_MC2_155 EVVSESGLGVP-DWVFSAIALIAVANGALLTMIMASRLAFGMAEHRLLPS
Mflv_4427|M.gilvum_PYR-GCK DVVAASGAAVP-DWLFSLIALIAVANGALLTMIMASRLTFGMSDQGLLPS
TH_4382|M.thermoresistible__bu EVVKAGAPGLPIEELLPWISMFAVSNTALINMLMASRLIYGMARQRVLPP
Mvan_5147|M.vanbaalenii_PYR-1 EVVRAGAPGLPIDAILPFMTMFAVSNTALINMLMASRLIYGMARQHVLPP
Mb3281c|M.bovis_AF2122/97 ATAFQANGVYWASGIISVGALAGLTTVVMVLMLGQCRVLFAMARDGLVPR
Rv3253c|M.tuberculosis_H37Rv ATAFQANGVYWASGIISVGALAGLTTVVMVLMLGQCRVLFAMARDGLVPR
MMAR_1290|M.marinum_M ATAFKANGVYWASGIISVGALAGLTTVVMVLMLGQCRVLFAMARDGLLPR
MUL_2597|M.ulcerans_Agy99 ATAFKANGVYWASGIISVGALAGLTTVVMVLMLGQCRVLFAMARDGLLPR
MAV_4216|M.avium_104 ATAFTANGIHWASKIISIGALAGLTTVVMVLMLGQCRVLFAMARDGLLPR
MAB_2598|M.abscessus_ATCC_1997 DAVTAAG-AGWMEPVVRIGAVVAATGSLLALILGVSRTTFAMARDRHLPH
MLBr_01974|M.leprae_Br4923 VKLAAAVGNNTFRTIIVVGALVSMFGINVAASFGAPRLWTALSDGRILPT
. :. :: . : : * .:: :*
MSMEG_6727|M.smegmatis_MC2_155 VLSRVLTKRRTPWVAIVATTAVAMVLTAIG-------ELSTLAETVVLLL
Mflv_4427|M.gilvum_PYR-GCK VFARVLPNRRTPWVAIVATTAVAMALTLVG-------DLSMLAETVVLLL
TH_4382|M.thermoresistible__bu VLGSVHPRRRTPWVAIIFTTLIAFGLIFYVTAFADSTAIAVLGGTTSLLL
Mvan_5147|M.vanbaalenii_PYR-1 VLGTVHPTRRSPWVAILFTTLIAFGLIIYVSTFASSNAIAVLGGTTSLLL
Mb3281c|M.bovis_AF2122/97 QLAKTGSRGTPVRVTVLVAVLVATTASVFP--------ITKLEEMVNVGT
Rv3253c|M.tuberculosis_H37Rv QLAKTGSRGTPVRVTVLVAVLVATTASVFP--------ITKLEEMVNVGT
MMAR_1290|M.marinum_M QLAKTGARGTPVRITVLVAVVVGATASVFP--------ITKLEEMVNVGT
MUL_2597|M.ulcerans_Agy99 QLAKTGARGTPVRITVLVAVVVGATASVFP--------ITKLEEMVNVGT
MAV_4216|M.avium_104 ALAKTGSRGTPVRITVLVALVVAVTASVFP--------ITKLEEMVNVGT
MAB_2598|M.abscessus_ATCC_1997 TLATVHPKYGVPYRAEILVGLVVSVLAATAD-------LRGAIGFSSFAV
MLBr_01974|M.leprae_Br4923 RLSSKN-RFGVPIVAFGITAFLTLAFPLSLR-----FDSLNLTGLAVIAR
:. : . : .
MSMEG_6727|M.smegmatis_MC2_155 LFVFTATNIAVLVLRRDTV-----EHDHFRVWTAVPVLGVASCVLLMTQQ
Mflv_4427|M.gilvum_PYR-GCK LVVFLSANVSVLVLRRDHV-----EQDHFRVWTFVPVLGIASCLLLLTQQ
TH_4382|M.thermoresistible__bu LAVFAMVNVAVLVLRRDESRDPVVARRHFRTPTFLPVVGFVASLYLVTPL
Mvan_5147|M.vanbaalenii_PYR-1 LAVFAVVNVAVLALRRDVR----EAGGHFKTPTALPVIGFLASLFLVTPL
Mb3281c|M.bovis_AF2122/97 LFAFILVSAGVVVLRRTRPD--LQRGFTAPWVPLLPIAAVCACLWLMLNL
Rv3253c|M.tuberculosis_H37Rv LFAFILVSAGVVVLRRTRPD--LQRGFTAPWVPLLPIAAVCACLWLMLNL
MMAR_1290|M.marinum_M LFAFVLVSAGVIVLRKKRPD--LERGFRAPWVPVLPIASLCACLWLMLNL
MUL_2597|M.ulcerans_Agy99 LFAFVLVSAGVIVLRKKRPD--LERGFRAPWVPVLPIASLCACLWLMLNL
MAV_4216|M.avium_104 LFAFVLVSAGVMVLRRTRPD--LERGFKAPWVPVLPIASIGACVWLMLNL
MAB_2598|M.abscessus_ATCC_1997 LVYYAVANASAWTLTPSQGR----------PARIIPAVGLVGCVIVSLAL
MLBr_01974|M.leprae_Br4923 FIQFIIVPIALFALARSRTTNSVYVKRNVFTDKVLPITAVVVSAGLMVSF
: : . . .* :* .. . :
MSMEG_6727|M.smegmatis_MC2_155 T---AKVWMFAGILMAVGVVLYFVAR-------AATNRTADETV------
Mflv_4427|M.gilvum_PYR-GCK T---AKVWLLAAALLAVGVVLYLLAKRFG---GQGVQDTGDAAPSRSE--
TH_4382|M.thermoresistible__bu SGRPAQQYLLSVVLIVIGVLLFGVTMAINRQLGIPTRAVGDPAKMVLPPD
Mvan_5147|M.vanbaalenii_PYR-1 SGRPAQQYIVAATLVAIGVVLFFVTQLINRQLGIRDGHITDPTRLGSGTD
Mb3281c|M.bovis_AF2122/97 T---ALTWIRFGIWLVAGTAIYVGYGRRHSAQGLRQARESATRRC-----
Rv3253c|M.tuberculosis_H37Rv T---ALTWIRFGIWLVAGTAIYVGYGRRHSAQGLRQARESATRRC-----
MMAR_1290|M.marinum_M T---ALTWIRFGVWLLVGTVIYVGYGRKHSMVGRRQLAEAAMDK------
MUL_2597|M.ulcerans_Agy99 T---ALTWIRFGVWLLVGTVIYVGYGRKHSMVGRRQLAEAAMDK------
MAV_4216|M.avium_104 T---ALTWVRFGIWLVVGTAIYAGYGYWHSVQGRRDAGEPDRSDALAADP
MAB_2598|M.abscessus_ATCC_1997 P---LSSVAAGTGVLAVGVIVYALRRVSGPGRRE----------------
MLBr_01974|M.leprae_Br4923 DYRSIFLTRGGPNYFSISLIAITFIGIPAVAYLHYYRTSGIK--------
. .
MSMEG_6727|M.smegmatis_MC2_155 -----
Mflv_4427|M.gilvum_PYR-GCK -----
TH_4382|M.thermoresistible__bu -----
Mvan_5147|M.vanbaalenii_PYR-1 -----
Mb3281c|M.bovis_AF2122/97 -----
Rv3253c|M.tuberculosis_H37Rv -----
MMAR_1290|M.marinum_M -----
MUL_2597|M.ulcerans_Agy99 -----
MAV_4216|M.avium_104 NSPVP
MAB_2598|M.abscessus_ATCC_1997 -----
MLBr_01974|M.leprae_Br4923 -----