For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSQQPGALRRELGTFDAVMIGLGSMIGAGIFAALAPAARSAGSALLIGLCVAAVIAYCNATSSARLAAV YPASGGTYVYGRERLGDFWGYLAGWGFVVGKTASCAAMALTIGFYAWPGHAHAVAVASVVALTAINYVGV QKSAWLTRVIVAIVLSVLAAIVATAAGSGSVDIAALDLTGTSWRGVLQAAGLLFFAFAGYARIATLGEEV RDPARTIPRAIPIALGIALVVYVVVALAVLITLGPSRLASSAAPLLDAVTAAGAGWMEPVVRIGAVVAAT GSLLALILGVSRTTFAMARDRHLPHTLATVHPKYGVPYRAEILVGLVVSVLAATADLRGAIGFSSFAVLV YYAVANASAWTLTPSQGRPARIIPAVGLVGCVIVSLALPLSSVAAGTGVLAVGVIVYALRRVSGPGRRE
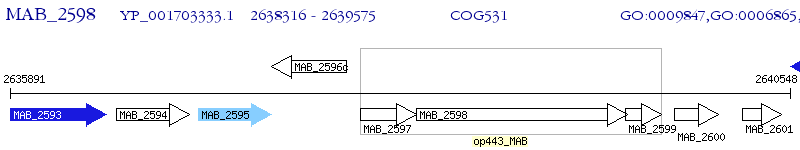
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2598 | - | - | 100% (419) | hypothetical protein MAB_2598 |
| M. abscessus ATCC 19977 | MAB_3599c | - | 8e-40 | 29.62% (449) | putative amino acid permease |
| M. abscessus ATCC 19977 | MAB_3842 | - | 8e-38 | 29.12% (443) | cationic amino acid transport integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2022c | - | 1e-169 | 74.33% (409) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_0187 | - | 1e-150 | 68.38% (408) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv1999c | - | 1e-169 | 74.33% (409) | integral membrane protein |
| M. leprae Br4923 | MLBr_01974 | - | 1e-20 | 23.91% (435) | putative permease |
| M. marinum M | MMAR_3621 | rocE | 8e-40 | 29.64% (442) | cationic amino acid transport integral membrane protein RocE |
| M. avium 104 | MAV_4216 | - | 4e-37 | 28.01% (457) | cationic amino acid transporter |
| M. smegmatis MC2 155 | MSMEG_0812 | - | 1e-162 | 72.37% (409) | amino acid transporter |
| M. thermoresistible (build 8) | TH_3126 | - | 5e-92 | 69.80% (255) | PUTATIVE putative amino acid permease-associated region |
| M. ulcerans Agy99 | MUL_1244 | rocE | 1e-40 | 29.41% (442) | cationic amino acid transport integral membrane protein RocE |
| M. vanbaalenii PYR-1 | Mvan_0718 | - | 1e-151 | 68.29% (410) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2022c|M.bovis_AF2122/97 --------------------------------------------------
Rv1999c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2598|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_0187|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0718|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0812|M.smegmatis_MC2_155 --------------------------------------------------
TH_3126|M.thermoresistible__bu --------------------------------------------------
MMAR_3621|M.marinum_M --------------------------------------------------
MUL_1244|M.ulcerans_Agy99 --------------------------------------------------
MAV_4216|M.avium_104 --------------------------------------------------
MLBr_01974|M.leprae_Br4923 MFLSADVSCIEIVDINCSELRNDRRIPPQLALFCTNAANKAESAVTSHGQ
Mb2022c|M.bovis_AF2122/97 --------------------------------------------MRRPLD
Rv1999c|M.tuberculosis_H37Rv --------------------------------------------MRRPLD
MAB_2598|M.abscessus_ATCC_1997 --------------------------------------------MSSQQ-
Mflv_0187|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0718|M.vanbaalenii_PYR-1 --------------------------------------------MSSPGG
MSMEG_0812|M.smegmatis_MC2_155 --------------------------------------------MSTPGG
TH_3126|M.thermoresistible__bu --------------------------------------------------
MMAR_3621|M.marinum_M ---------------------------MPATSIRLREQMLRRRPVGGAPV
MUL_1244|M.ulcerans_Agy99 ---------------------------MPATSIRLREQMLRRRPAGGAPV
MAV_4216|M.avium_104 ------------------------------MASRWRTKSVEQSIAD----
MLBr_01974|M.leprae_Br4923 LPIMANRRWAPHTRRQSGRATSTCSSLMRFSASESRAHSRFECYTLVRWS
Mb2022c|M.bovis_AF2122/97 PRDIPDELRRRLGLLDAVVIGLGSMIGAGIFAALAPAAYAAG-SGLLLGL
Rv1999c|M.tuberculosis_H37Rv PRDIPDELRRRLGLLDAVVIGLGSMIGAGIFAALAPAAYAAG-SGLLLGL
MAB_2598|M.abscessus_ATCC_1997 ----PGALRRELGTFDAVMIGLGSMIGAGIFAALAPAARSAG-SALLIGL
Mflv_0187|M.gilvum_PYR-GCK -------MRRRLGTFDTVTLGLGSMIGAGIFVALAPAAAAAG-TGLLVGL
Mvan_0718|M.vanbaalenii_PYR-1 S-----TLDRRLGVFDTVTIGLGSMIGAGIFVALAPAAAAAG-SGLLIGL
MSMEG_0812|M.smegmatis_MC2_155 -------LQRRLGTFDAVTIGLGSMIGAGIFAAIGPAAAAAG-SGLLIGL
TH_3126|M.thermoresistible__bu -------LPRRLGTLDAVTIGLGAMIGAGIFVALGPAAAAAG-PWLLAGL
MMAR_3621|M.marinum_M AEGAAGSLKRSFGTFELTMFGVGSTVGTGIFFVLAEAVPEAG-PAVIVSF
MUL_1244|M.ulcerans_Agy99 AEGAAGSLKRSFGTFELTMFGVGSTVGTGIFFVLAEAVPEAG-PAVIVSF
MAV_4216|M.avium_104 TDEPDTRLRKDLTWWDLVVFGVAVVIGAGIFTVTASTTGDITGPAIWVSF
MLBr_01974|M.leprae_Br4923 IRGQGGRAISKLGFWSVVMLGINSIIGAGIFMTPGEVIKIAG-PFAPIAY
: . . :*: :*:*** . . . . .
Mb2022c|M.bovis_AF2122/97 AVAAVVAYCNAISSARLAARYPASGGTYVYGRMRLGDFWGYLAGWGFVVG
Rv1999c|M.tuberculosis_H37Rv AVAAVVAYCNAISSARLAARYPASGGTYVYGRMRLGDFWGYLAGWGFVVG
MAB_2598|M.abscessus_ATCC_1997 CVAAVIAYCNATSSARLAAVYPASGGTYVYGRERLGDFWGYLAGWGFVVG
Mflv_0187|M.gilvum_PYR-GCK AVAALIAVCNAMSSARLAARYPESGGTYVYGRERLGPFWGHTAGWSFIVG
Mvan_0718|M.vanbaalenii_PYR-1 AVAAVVAVCNAMSSARLAARYPQSGGTYVYGRERLGPFWGHAAGWSFIVG
MSMEG_0812|M.smegmatis_MC2_155 ALAAVVAYCNATSSARLAAIYPQSGGTYVYGRERLGEFWGYLAGWSFAVG
TH_3126|M.thermoresistible__bu GIAAAVAYCNATASARLAARYPQSGGTYVYGRERLGAFWGYTAGFSFVVG
MMAR_3621|M.marinum_M IIAGVAAGLAAICYAELASAVPVSGSSYSYAYATLGEAVAMGVASCLLLE
MUL_1244|M.ulcerans_Agy99 IIAGVAAGLAAICCAELASAVPVSGSSYSYAYATLGEAVAMGVASCLLLE
MAV_4216|M.avium_104 VIAAGTCALAALCYAEFASTLPVAGSAYTFSYATFGEFLAWIIGWNLLLE
MLBr_01974|M.leprae_Br4923 TLAGIFAAVLAIVFATAAKYVKTNGSSYAYTTAAFGHRIGIYVGVTHAIT
:*. . * * * *.:* : :* . . :
Mb2022c|M.bovis_AF2122/97 KTASCAAMALTVGFYVWP---------------------------AQAHA
Rv1999c|M.tuberculosis_H37Rv KTASCAAMALTVGFYVWP---------------------------AQAHA
MAB_2598|M.abscessus_ATCC_1997 KTASCAAMALTIGFYAWP---------------------------GHAHA
Mflv_0187|M.gilvum_PYR-GCK KTASCAAMALTVGYYVWP---------------------------AYANA
Mvan_0718|M.vanbaalenii_PYR-1 KTASCAAMALTVGFYAWP---------------------------EYANA
MSMEG_0812|M.smegmatis_MC2_155 KTASCAAMALTVGSYAWP---------------------------EHANA
TH_3126|M.thermoresistible__bu KTASCAAMALTIGFYLWP---------------------------GHAHA
MMAR_3621|M.marinum_M YGVATAAVAVGWSGYVNKLLHNVFGFQLPQALSEAPWDERPGEAAGYLNL
MUL_1244|M.ulcerans_Agy99 YGVATAAVAVGLSGYVNKLLHNVFGFQLPQALSEAPWDERPGEAAGYLNL
MAV_4216|M.avium_104 LAIGAAVVAKGWSSYLG----TVFGFSG----------GTVKFGAAQLDW
MLBr_01974|M.leprae_Br4923 ASIAWGVLASSFVSTLLRVAFPEKIWAD------------NDELISVKTL
. ..:*
Mb2022c|M.bovis_AF2122/97 VAVAVVVALTAVNYAGIQKSAWLTRSIVAVVLVVLTAVVVAAYGSGAADP
Rv1999c|M.tuberculosis_H37Rv VAVAVVVALTAVNYAGIQKSAWLTRSIVAVVLVVLTAVVVAAYGSGAADP
MAB_2598|M.abscessus_ATCC_1997 VAVASVVALTAINYVGVQKSAWLTRVIVAIVLSVLAAIVATAAGSGSVDI
Mflv_0187|M.gilvum_PYR-GCK VAVGSVVALTAVSYAGVQRSATLTRVIVALVLGVLAMVVVVVFGSGDVDA
Mvan_0718|M.vanbaalenii_PYR-1 VAVASVVALTAVGYAGVQRSAALTRIIVALVLAVLAMVVVVLLGFGGADT
MSMEG_0812|M.smegmatis_MC2_155 VAVGAVVALTAVNYVGVQKSAVLTRVIVAVVLAILATVVVLLLGSGDADP
TH_3126|M.thermoresistible__bu IAVAAVVALTALTYLGIEKSALLTRLVVAGVLGVLTAVVVLVLGFGAPAG
MMAR_3621|M.marinum_M PAIILIGLCALLLIRGVSESAKVNAIMVLIKLGVLGMFVILALTAYDTNH
MUL_1244|M.ulcerans_Agy99 PAIILIGLCALLLIRGVSESAKVNAIMVLIKLGVLGMFVILALTAYDTNH
MAV_4216|M.avium_104 GALVIVGGVATLIALGTKLSSRFSAVITGIKVSVVVFVVVVGVFYIKRAN
MLBr_01974|M.leprae_Br4923 TFLVFIAVLLAINIFGNLVIRWANGISALGKIFALSVFIAGGLWTIITQH
: : : * . . : : .:
Mb2022c|M.bovis_AF2122/97 ARLDIG------------------------VDAHVWGMLQAAGLLFFAFA
Rv1999c|M.tuberculosis_H37Rv ARLDIG------------------------VDAHVWGMLQAAGLLFFAFA
MAB_2598|M.abscessus_ATCC_1997 AALDL-------------------------TGTSWRGVLQAAGLLFFAFA
Mflv_0187|M.gilvum_PYR-GCK SRLALG------------------------PDVDAYGVLQAAGLLFFAFA
Mvan_0718|M.vanbaalenii_PYR-1 SRLALG------------------------DDVTVYGVLQAAGLLFFAFA
MSMEG_0812|M.smegmatis_MC2_155 ARLSLG------------------------DDVSAVGVLQAAGLLFFAFA
TH_3126|M.thermoresistible__bu RVAGGG------------------------TDIGAGAVLQSAALLFFAFA
MMAR_3621|M.marinum_M LKDFAP--------------------------FGVAGVGAAAGTIFFSFI
MUL_1244|M.ulcerans_Agy99 LKDFAP--------------------------FGVAGVGAAAGTIFFSFI
MAV_4216|M.avium_104 YSPFIPKPEAGGQAKGIDQSVLSLLTGAHTSHYGWYGVLAGASIVFFAFI
MLBr_01974|M.leprae_Br4923 VNNYVTSRSAYQPTTYSLLG------VTEIGKSTAASMTLATIVALYAFT
.: .: :::*
Mb2022c|M.bovis_AF2122/97 GYARIATLGEEVRDPARTIPRAIPLALGITLAVYALVAVAVIAVLGPQRL
Rv1999c|M.tuberculosis_H37Rv GYARIATLGEEVRDPARTIPRAIPLALGITLAVYALVAVAVIAVLGPQRL
MAB_2598|M.abscessus_ATCC_1997 GYARIATLGEEVRDPARTIPRAIPIALGIALVVYVVVALAVLITLGPSRL
Mflv_0187|M.gilvum_PYR-GCK GYARIATLGEEVRDPARTIPRAIGLALVIALLTYAVVAVAVLAQLGSTGL
Mvan_0718|M.vanbaalenii_PYR-1 GYARIATLGEEVRDPARTIPRAVALALVIALMVYAVVAVAVVSELGVAGL
MSMEG_0812|M.smegmatis_MC2_155 GYARIATLGEEVRDPARIIPRAIRIALGLTLVVYAVVAGAVLAVLGGTDL
TH_3126|M.thermoresistible__bu GYARIATLGEEVRDPARTIRRAIPLALGITLTVYGAVTAAVLVALGSAGL
MMAR_3621|M.marinum_M GLDAVSTAGEEVRNPQQSVPRALIAALFVVTGVYVLVAFAALGTQPWQMF
MUL_1244|M.ulcerans_Agy99 GLDAVSTAGEEVRNPQQSVPRALIAALFVVTGVYVLVAFAALGTQPWQMF
MAV_4216|M.avium_104 GFDIVATMAEETKHPQRDVPRGILASLGIVTLLYVAVAVVLSGMVSYTQL
MLBr_01974|M.leprae_Br4923 GFESIANAAEEMNTPDRTLPKAIPLTILTVGVIYILALVVAMLLGSDKIV
* ::. .** . * : : :.: :: . * . . .
Mb2022c|M.bovis_AF2122/97 AR----AAAPLSEAMRVAGVNWLIPVVQIGAAVAALGSLLALILGVSRTT
Rv1999c|M.tuberculosis_H37Rv AR----AAAPLSEAMRVAGVNWLIPVVQIGAAVAALGSLLALILGVSRTT
MAB_2598|M.abscessus_ATCC_1997 AS----SAAPLLDAVTAAGAGWMEPVVRIGAVVAATGSLLALILGVSRTT
Mflv_0187|M.gilvum_PYR-GCK AT----ADAPLADAVRAAGLPALAPVVAVGAALAALGALLALLLGVSRTT
Mvan_0718|M.vanbaalenii_PYR-1 AS----ATAPLSDAVRAAGFPGLTPVVNVGAAIAALGALLALLLGVSRTT
MSMEG_0812|M.smegmatis_MC2_155 AG----ATAPLADAVAAVGAGQWQPLVRAGAAIAALGSLLALILGVSRTT
TH_3126|M.thermoresistible__bu AR----SPAPVADAVTAAXX---------HGRQQPFGELPG---------
MMAR_3621|M.marinum_M AGQEEAGLATILDNVTHS--AWASTVLAMGAVISIFTVTLVTMYGQTRIL
MUL_1244|M.ulcerans_Agy99 AGQEEAGLATILDNVTHS--AWASTVLAMGAVISIFTVTLVTMYGQTRIL
MAV_4216|M.avium_104 KTMPGRGQANLATAFTANGIHWASKIISIGALAGLTTVVMVLMLGQCRVL
MLBr_01974|M.leprae_Br4923 AT------NHTVKLAAAVGNNTFRTIIVVGALVSMFGINVAASFGAPRLW
.
Mb2022c|M.bovis_AF2122/97 LAMARDRHLPRWLAAVHPRFKVPFRAELVVGAVVAALAATADIRGA--IG
Rv1999c|M.tuberculosis_H37Rv LAMARDRHLPRWLAAVHPRFKVPFRAELVVGAVVAALAATADIRGA--IG
MAB_2598|M.abscessus_ATCC_1997 FAMARDRHLPHTLATVHPKYGVPYRAEILVGLVVSVLAATADLRGA--IG
Mflv_0187|M.gilvum_PYR-GCK LAMARDRHLPHPLAHVHPGHGTPQRAEVAVGVVVAVVAAVVDVRSA--IG
Mvan_0718|M.vanbaalenii_PYR-1 LAMARDRYLPAGLAAVHPKHGTPHHAEVAVGLVVAVVAAFVDLRGA--IG
MSMEG_0812|M.smegmatis_MC2_155 LAMARDHHLPSALGAVHPRFGVPHRAEVTVGVVVAVLAAVADLRGA--IG
TH_3126|M.thermoresistible__bu -AEEVDRGHPQRIPDAR---RHPGHVEHRS------SPTTCAPR------
MMAR_3621|M.marinum_M FAMGRDGLLPARFAAVHPRFQTPVSNTVVVAIAAAVLAALVPLNRL--AD
MUL_1244|M.ulcerans_Agy99 FAMGRDGLLPARFAAVHPRFQTPVSNTVVVAIAAAVLAALVPLNRL--AD
MAV_4216|M.avium_104 FAMARDGLLPRALAKTGSR-GTPVRITVLVALVVAVTASVFPITKL--EE
MLBr_01974|M.leprae_Br4923 TALSDGRILPTRLSSKN-RFGVPIVAFGITAFLTLAFPLSLRFDSLNLTG
* . * : * .
Mb2022c|M.bovis_AF2122/97 FSSFGVLVYYAIANASALTLGLDEGRPRR----------LIPLVGLIGCV
Rv1999c|M.tuberculosis_H37Rv FSSFGVLVYYAIANASALTLGLDEGRPRR----------LIPLVGLIGCV
MAB_2598|M.abscessus_ATCC_1997 FSSFAVLVYYAVANASAWTLTPSQGRPAR----------IIPAVGLVGCV
Mflv_0187|M.gilvum_PYR-GCK FSSFGVLLYYAVANASAWTLTATPR--AR----------LVPAVGLIGCV
Mvan_0718|M.vanbaalenii_PYR-1 FSSFGVLLYYAIANASAWTLG------AR----------LIPAIGLVGCL
MSMEG_0812|M.smegmatis_MC2_155 FSSFAVLLYYAIANASAATLG------GR----------LVPAVGLVGCL
TH_3126|M.thermoresistible__bu -----------------------------------------PLTWRTTWA
MMAR_3621|M.marinum_M MVSIGTLTAFIVVSVGVIILRVREPALPRGFWV--PGYPVTPILSILACG
MUL_1244|M.ulcerans_Agy99 MVSIGTLTAFIVVSVGVIILRVREPALPRGFWV--PGYPVTPILSILACG
MAV_4216|M.avium_104 MVNVGTLFAFVLVSAGVMVLRRTRPDLERGFKA--PWVPVLPIASIGACV
MLBr_01974|M.leprae_Br4923 LAVIARFIQFIIVPIALFALARSRTTNSVYVKRNVFTDKVLPITAVVVSA
*
Mb2022c|M.bovis_AF2122/97 VLAFALPLSSVAA---GAAVLGVGVAAYGVRRIITRRARQTDSGDTQRSG
Rv1999c|M.tuberculosis_H37Rv VLAFALPLSSVAA---GAAVLGVGVAAYGVRRIITRRARQTDSGDTQRSG
MAB_2598|M.abscessus_ATCC_1997 IVSLALPLSSVAA---GTGVLAVGVIVYALRRVSGPGRRE----------
Mflv_0187|M.gilvum_PYR-GCK VLAFTLPPVSVLA---GAAVVLVGMLAYTTGGALRRGV------------
Mvan_0718|M.vanbaalenii_PYR-1 VLAFTLPLTSVLA---GAAVVLVGMIAYTIGGTR----------------
MSMEG_0812|M.smegmatis_MC2_155 VLAFTLPLSSVLA---GLAVVAAGAVVYGVRRIS----------------
TH_3126|M.thermoresistible__bu PMPDPQPVTTTRR---PS--------------------------------
MMAR_3621|M.marinum_M YLLYSLHWYTWIA---FGGWVGIALIFYLFWGRHHSRLN---------AP
MUL_1244|M.ulcerans_Agy99 YLLYSLHWYTWIA---FGGWVGIALIFYLFWGRHHSRLN---------AP
MAV_4216|M.avium_104 WLMLNLTALTWVR---FGIWLVVGTAIYAGYGYWHSVQGRRDAGEPDRSD
MLBr_01974|M.leprae_Br4923 GLMVSFDYRSIFLTRGGPNYFSISLIAITFIGIPAVAYLHYYRTSGIK--
: :
Mb2022c|M.bovis_AF2122/97 HPSAT------
Rv1999c|M.tuberculosis_H37Rv HPSAT------
MAB_2598|M.abscessus_ATCC_1997 -----------
Mflv_0187|M.gilvum_PYR-GCK -----------
Mvan_0718|M.vanbaalenii_PYR-1 -----------
MSMEG_0812|M.smegmatis_MC2_155 -----------
TH_3126|M.thermoresistible__bu -----------
MMAR_3621|M.marinum_M ALHDNPTGEER
MUL_1244|M.ulcerans_Agy99 ALHDNPTGEER
MAV_4216|M.avium_104 ALAADPNSPVP
MLBr_01974|M.leprae_Br4923 -----------