For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAGRRRMKSVEQSIADTDEPTTRLRKDLTWWDLVVFGVSVVIGAGIFTVTASTAGDITGPAIWISFLIAA ATCALAALCYAEFASTLPVAGSAYTFSYATFGEFLAWVIGWNLVLELAMGAAVVAKGWSSYLGTVFGFGN GTGHLGSLQLDWGALVIVTLVATLIALGTKLSSRFSAVVTAIKVSVVVLVVVVGAFYIRAANYSPFIPEP EVQHHGGGLDQSVFSLLTGAQGSHYGWYGVLAGASIVFFAFIGFDIVATMAEETKRPQRDVPRGILASLG VVTLLYVAVSVVLSGMVPYTQLRTVPGRGPANLATAFQANGVYWASGIISVGALAGLTTVVMVLMLGQCR VLFAMARDGLVPRQLAKTGSRGTPVRVTVLVAVLVATTASVFPITKLEEMVNVGTLFAFILVSAGVVVLR RTRPDLQRGFTAPWVPLLPIAAVCACLWLMLNLTALTWIRFGIWLVAGTAIYVGYGRRHSAQGLRQARES ATRRC
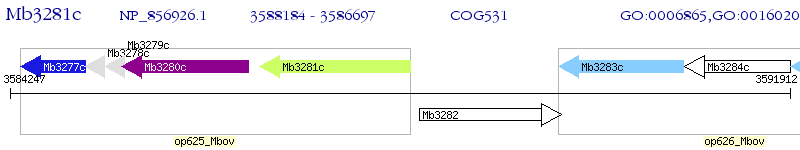
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3281c | - | - | 100% (495) | cationic amino acid transport integral membrane protein |
| M. bovis AF2122 / 97 | Mb2347c | rocE | 4e-79 | 35.88% (471) | cationic amino acid transport integral membrane protein RocE |
| M. bovis AF2122 / 97 | Mb2022c | - | 2e-32 | 26.36% (478) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4722 | - | 0.0 | 72.65% (490) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv3253c | - | 0.0 | 100.00% (495) | cationic amino acid transport integral membrane protein |
| M. leprae Br4923 | MLBr_01974 | - | 4e-20 | 23.34% (437) | putative permease |
| M. abscessus ATCC 19977 | MAB_3599c | - | 0.0 | 67.63% (485) | putative amino acid permease |
| M. marinum M | MMAR_1290 | - | 0.0 | 86.15% (491) | cationic amino acid transport integral membrane protein |
| M. avium 104 | MAV_4216 | - | 0.0 | 84.87% (489) | cationic amino acid transporter |
| M. smegmatis MC2 155 | MSMEG_1838 | - | 0.0 | 73.33% (495) | cationic amino acid transporter |
| M. thermoresistible (build 8) | TH_0471 | - | 0.0 | 70.85% (494) | POSSIBLE CATIONIC AMINO ACID TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_2597 | - | 0.0 | 85.74% (491) | cationic amino acid transport integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_1740 | - | 0.0 | 74.17% (484) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3281c|M.bovis_AF2122/97 --------------------------------------------------
Rv3253c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1290|M.marinum_M --------------------------------------------------
MUL_2597|M.ulcerans_Agy99 --------------------------------------------------
MAV_4216|M.avium_104 --------------------------------------------------
Mflv_4722|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1740|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1838|M.smegmatis_MC2_155 --------------------------------------------------
TH_0471|M.thermoresistible__bu --------------------------------------------------
MAB_3599c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01974|M.leprae_Br4923 MFLSADVSCIEIVDINCSELRNDRRIPPQLALFCTNAANKAESAVTSHGQ
Mb3281c|M.bovis_AF2122/97 ---MAG----------RRRMKSVEQSIADTDEPTTRLR------------
Rv3253c|M.tuberculosis_H37Rv ---MAG----------RRRMKSVEQSIADTDEPTTRLR------------
MMAR_1290|M.marinum_M ---MAG----------RLRTKSVEQSIADTDEPETRLR------------
MUL_2597|M.ulcerans_Agy99 ---MAG----------GLRTKSVEQSIADTDEPETRLR------------
MAV_4216|M.avium_104 ---MAS----------RWRTKSVEQSIADTDEPDTRLR------------
Mflv_4722|M.gilvum_PYR-GCK ---MTQEPVTQAP--GRLRLKSVEQSIADTDEPGTKLR------------
Mvan_1740|M.vanbaalenii_PYR-1 ---MVQEPGAQTV--GRRRTKSVEQSIADTDEPSTRLR------------
MSMEG_1838|M.smegmatis_MC2_155 ---MTT----------RWRTKSVEQSIADTDEPSTRLR------------
TH_0471|M.thermoresistible__bu ---MTD----------RLRVKSVEQSIADTDEPDSRLR------------
MAB_3599c|M.abscessus_ATCC_199 ---MPGS--------GLWRTKSVEQSIADTDEPETKLR------------
MLBr_01974|M.leprae_Br4923 LPIMANRRWAPHTRRQSGRATSTCSSLMRFSASESRAHSRFECYTLVRWS
* * .*. .*: . . :: :
Mb3281c|M.bovis_AF2122/97 ---------KDLTWWDLVVFGVSVVIGAGIFTVTASTAGDITGPAIWISF
Rv3253c|M.tuberculosis_H37Rv ---------KDLTWWDLVVFGVSVVIGAGIFTVTASTAGDITGPAIWISF
MMAR_1290|M.marinum_M ---------KDLTWRDLVVFGVSVVIGAGIFTVTASTAGDITGPAIWISF
MUL_2597|M.ulcerans_Agy99 ---------KDLTWRDLVVFGVSVVIGAGIFTVTASTAGDITGPAIWISF
MAV_4216|M.avium_104 ---------KDLTWWDLVVFGVAVVIGAGIFTVTASTTGDITGPAIWVSF
Mflv_4722|M.gilvum_PYR-GCK ---------KDLNWWDLTVFGVSVVIGAGIFTITASTAANITGPAISLSF
Mvan_1740|M.vanbaalenii_PYR-1 ---------KELNWWDLTVFGVSVVIGAGIFTITASTAANITGPAISLSF
MSMEG_1838|M.smegmatis_MC2_155 ---------KDLTWWDLTVFGVSVVIGAGIFTITASTAGNLTGPAISISF
TH_0471|M.thermoresistible__bu ---------KDLSWWDLTVFGVSVVIGAGIFTVTASTAGNITGPAISVSF
MAB_3599c|M.abscessus_ATCC_199 ---------RDLTWWDLTVFGVSVVIGAGIFTVTASTAANVTGPAISLSF
MLBr_01974|M.leprae_Br4923 IRGQGGRAISKLGFWSVVMLGINSIIGAGIFMTPG-EVIKIAGPFAPIAY
.* : .:.::*: :****** .. . .::** :::
Mb3281c|M.bovis_AF2122/97 LIAAATCALAALCYAEFASTLPVAGSAYTFSYATFGEFLAWVIGWNLVLE
Rv3253c|M.tuberculosis_H37Rv LIAAATCALAALCYAEFASTLPVAGSAYTFSYATFGEFLAWVIGWNLVLE
MMAR_1290|M.marinum_M LIAAGTCALAALCYAEFASTLPVAGSAYTFSYATFGEFLAWVIGWNLVLE
MUL_2597|M.ulcerans_Agy99 LIAAGTCALAALCYAEFASTLPVAGSAYTFSYATFGEFLAWVIGWNLVLE
MAV_4216|M.avium_104 VIAAGTCALAALCYAEFASTLPVAGSAYTFSYATFGEFLAWIIGWNLLLE
Mflv_4722|M.gilvum_PYR-GCK IFAAIACGLAALCYAEFASTVPVAGSAYTFSYATFGEFVAWIIGWDLILE
Mvan_1740|M.vanbaalenii_PYR-1 IIAAIACGLAALCYAEFASTVPVAGSAYTFSYATFGEFVAWIIGWDLILE
MSMEG_1838|M.smegmatis_MC2_155 LIAAVACGLAALCYAEFASTVPVAGSAYTFSYATFGEFVAWIIGWDLILE
TH_0471|M.thermoresistible__bu IIAAIACGLAALCYAEFASTVPVAGSAYTFSYATFGEFVAWIIGWDLILE
MAB_3599c|M.abscessus_ATCC_199 IMAAIGCGLAAMCYAEFASTIPVAGSAYTFSYATFGEFAAWILGWDLILE
MLBr_01974|M.leprae_Br4923 TLAGIFAAVLAIVFATAAKYVKTNGSSYAYTTAAFGHRIGIYVGVTHAIT
:*. ..: *: :* *. : . **:*::: *:**. . :* :
Mb3281c|M.bovis_AF2122/97 LAMGAAVVAKGWSSYLGTVFGFGNGTGHLGS-LQLDWGALVIVTLVATLI
Rv3253c|M.tuberculosis_H37Rv LAMGAAVVAKGWSSYLGTVFGFGNGTGHLGS-LQLDWGALVIVTLVATLI
MMAR_1290|M.marinum_M LAIGAAVVAKGWSSYLGTVFGFSGGTADFGS-IRLDWGALLIVSIVSLLV
MUL_2597|M.ulcerans_Agy99 LAIGAAVVAKGWSSYLGTVFGFSGGTADFGS-IRLDWGALLSVSIVSLLV
MAV_4216|M.avium_104 LAIGAAVVAKGWSSYLGTVFGFSGGTVKFGA-AQLDWGALVIVGGVATLI
Mflv_4722|M.gilvum_PYR-GCK FAVAAAVVAKGWSSYLGTVFGFAGGTAEVAG-FDLDWGALVIIAIVTAIL
Mvan_1740|M.vanbaalenii_PYR-1 FAVAAAVVAKGWSSYLGTVFGFAGGTTEVAG-FQLDWGALVIIALVTLIL
MSMEG_1838|M.smegmatis_MC2_155 FAVAAAVVAKGWSSYLGTVFNFGGGTMQIGGGLTLDWGALLIIAVVTFLL
TH_0471|M.thermoresistible__bu FSVAAAVVAKGWSSYLGEVFEFAGATTRLGP-ITLDWGALLIIGFVTVLL
MAB_3599c|M.abscessus_ATCC_199 FSVGAATVAKGWSSYLSTVFGLSSGAVHLGA-VKIDWGALLIVSVLTVLL
MLBr_01974|M.leprae_Br4923 ASIAWGVLASSFVSTLLRVAFPEKIWADNDELISVKTLTFLVFIAVLLAI
::. ..:*..: * * * :. ::: . : :
Mb3281c|M.bovis_AF2122/97 ALGTKLSSRFSAVVTAIKVSVVVLVVVVGAFYIRAANYSPFIPEPEVQHH
Rv3253c|M.tuberculosis_H37Rv ALGTKLSSRFSAVVTAIKVSVVVLVVVVGAFYIRAANYSPFIPEPEVQHH
MMAR_1290|M.marinum_M AYGTKLSSRFSAVVTTIKVAVVILVVVVGAFYVKAANYSPFIPEPEAGPE
MUL_2597|M.ulcerans_Agy99 AYGTKLSSRFSAVVTTIKVAVVILVVVVGAFYVKAANYSPFIPEPEAGPE
MAV_4216|M.avium_104 ALGTKLSSRFSAVITGIKVSVVVFVVVVGVFYIKRANYSPFIPKPEAGGQ
Mflv_4722|M.gilvum_PYR-GCK AMGTKLSASVSLAITALKVSVVLLVVIVGAFYIKMENFSPFIPAPEPGEA
Mvan_1740|M.vanbaalenii_PYR-1 VMGTKLSAGVSLAITAVKVSVVLLVVIVGAFYIKAENFSPFVPAPEEGGS
MSMEG_1838|M.smegmatis_MC2_155 ARGTKISAMVSLVITVIKVSVVLLVVFVGAFYINTANYTPFIPPAESGDG
TH_0471|M.thermoresistible__bu ATGTKLSANVSLAITVIKVGVVLLVVIVGAFFIKAANYTPFIPPAEAGGE
MAB_3599c|M.abscessus_ATCC_199 AVGTKLSSRVSLVITTIKVLVVLLVIIVGAFFIKTANYTPFIPPAEAGSS
MLBr_01974|M.leprae_Br4923 NIFGNLVIRWANGISALGKIFALSVFIAGGLWTIITQHVNNYVTSR----
:: : :: : ..: *...* :: :. ..
Mb3281c|M.bovis_AF2122/97 ---GGGLDQSVFSLLTGAQGSHYGWYGVLAGASIVFFAFIGFDIVATMAE
Rv3253c|M.tuberculosis_H37Rv ---GGGLDQSVFSLLTGAQGSHYGWYGVLAGASIVFFAFIGFDIVATMAE
MMAR_1290|M.marinum_M ---GRGADQSLLSLLTGAPGSHYGWYGVLAGASIVFFAFIGFDIVATMAE
MUL_2597|M.ulcerans_Agy99 ---GRGADQSLLSLLTGAPGSHYGWYGVLAGASIVFFAFIGFDIVATMAE
MAV_4216|M.avium_104 ---AKGIDQSVLSLLTGAHTSHYGWYGVLAGASIVFFAFIGFDIVATMAE
Mflv_4722|M.gilvum_PYR-GCK --GGTGAEQSLFSLMTGAEGSHYGWYGLLAGASIVFFAFIGFDIVATTAE
Mvan_1740|M.vanbaalenii_PYR-1 --GGTGTEQSLFSLLTGAEGSHYGWYGVLAGASIVFFAFIGFDIVATTAE
MSMEG_1838|M.smegmatis_MC2_155 SGGGTGAEQSLFSLLTGAEGSHYGWYGLLAGASIVFFAFIGFDIVATTAE
TH_0471|M.thermoresistible__bu --AGRSLDQSLFSLITGAEGSSYGWYGLLAGASIVFFAFIGFDIIATTAE
MAB_3599c|M.abscessus_ATCC_199 --SRQGLDQSLFSFVAGGSGSQYGWFGVLAGASIVFFAFIGFDVVATTAE
MLBr_01974|M.leprae_Br4923 ----SAYQPTTYSLLGVTEIGKSTAASMTLATIVALYAFTGFESIANAAE
. : : *:: . .: .: :.::** **: :*. **
Mb3281c|M.bovis_AF2122/97 ETKRPQRDVPRGILASLGVVTLLYVAVSVVLSGMVPYTQLRTVPGRGPAN
Rv3253c|M.tuberculosis_H37Rv ETKRPQRDVPRGILASLGVVTLLYVAVSVVLSGMVPYTQLRTVPGRGPAN
MMAR_1290|M.marinum_M ETKHPQRDVPRGILVSLGVVTLLYVAVSIVLSGMVSYTQLRTVSGGGHAN
MUL_2597|M.ulcerans_Agy99 ETKHPQRDVPRGILVSLGVVTLLYVAVSIVLSGMVSYTQLRTVSGGGHAN
MAV_4216|M.avium_104 ETKHPQRDVPRGILASLGIVTLLYVAVAVVLSGMVSYTQLKTMPGRGQAN
Mflv_4722|M.gilvum_PYR-GCK ETKVPQRDVPRGILASLGIVTVLYVAVAIVLSGMVSYKDLR---DAESPN
Mvan_1740|M.vanbaalenii_PYR-1 ETRNPQRDVPRGILASLAIVTVLYVAVAVVLTGMVSYTELR---DAETQN
MSMEG_1838|M.smegmatis_MC2_155 ETRNPQRDVPRGILGSLAIVTVLYVAVSVVLSGMVSYRDLKTV-DGESAN
TH_0471|M.thermoresistible__bu ETKNPQRDVARGIMATLGIVTVLYVAVSLVLTGMVHYTELR-A-AGENAN
MAB_3599c|M.abscessus_ATCC_199 ETRNPQKDVPRGIIASLVIVTVLYVAVTVVLSGMVKYSDLR--VADSHPN
MLBr_01974|M.leprae_Br4923 EMNTPDRTLPKAIPLTILTVGVIYILALVVAMLLGSDKIVA---TNHTVK
* . *:: :.:.* :: * ::*: . :* : : :
Mb3281c|M.bovis_AF2122/97 LATAFQANGVYWASGIISVGALAGLTTVVMVLMLGQCRVLFAMARDGLVP
Rv3253c|M.tuberculosis_H37Rv LATAFQANGVYWASGIISVGALAGLTTVVMVLMLGQCRVLFAMARDGLVP
MMAR_1290|M.marinum_M LATAFKANGVYWASGIISVGALAGLTTVVMVLMLGQCRVLFAMARDGLLP
MUL_2597|M.ulcerans_Agy99 LATAFKANGVYWASGIISVGALAGLTTVVMVLMLGQCRVLFAMARDGLLP
MAV_4216|M.avium_104 LATAFTANGIHWASKIISIGALAGLTTVVMVLMLGQCRVLFAMARDGLLP
Mflv_4722|M.gilvum_PYR-GCK LATAFSLNGVDWAAKVISIGALAGLTTVVIVLVLGQTRVLFAMSRDGLLP
Mvan_1740|M.vanbaalenii_PYR-1 LATAFSLNGVDWAAKVISIGALAGLTTVVIVLVLGQTRVLFAMSRDGLLP
MSMEG_1838|M.smegmatis_MC2_155 LATAFAQNGVDWAAKVISIGALAGLTTVVIVLMLGQTRVLFAMSRDGLMP
TH_0471|M.thermoresistible__bu LATAFAANGIDWAAKVISIGALAGLTTVVIVLVLGQTRVLFAMSRDGLLP
MAB_3599c|M.abscessus_ATCC_199 LSTAFHLNGVDWAAKVIAVGALAGLTTVVMVLMLGQIRVIFAMCRDGLLP
MLBr_01974|M.leprae_Br4923 LAAAVGNN---TFRTIIVVGALVSMFGINVAASFGAPRLWTALSDGRILP
*::*. * :* :***..: : :. :* *: *:. . ::*
Mb3281c|M.bovis_AF2122/97 RQLAKTGSRGTPVRVTVLVAVLVATTASVFPIT--KLEEMVNVGTLFAFI
Rv3253c|M.tuberculosis_H37Rv RQLAKTGSRGTPVRVTVLVAVLVATTASVFPIT--KLEEMVNVGTLFAFI
MMAR_1290|M.marinum_M RQLAKTGARGTPVRITVLVAVVVGATASVFPIT--KLEEMVNVGTLFAFV
MUL_2597|M.ulcerans_Agy99 RQLAKTGARGTPVRITVLVAVVVGATASVFPIT--KLEEMVNVGTLFAFV
MAV_4216|M.avium_104 RALAKTGSRGTPVRITVLVALVVAVTASVFPIT--KLEEMVNVGTLFAFV
Mflv_4722|M.gilvum_PYR-GCK RQLAKTGEHGTPVRITLLVGAVVAVTATVFPID--KLEEMVNIGTLFAFV
Mvan_1740|M.vanbaalenii_PYR-1 RQLAKTGEHGTPVRITLIVGAVVAVTATVFPIG--KLEEMVNIGTLFAFV
MSMEG_1838|M.smegmatis_MC2_155 RQLAKTGPHGTPVRTTLIVGGAVAVAASVFPIG--KLEEMVNIGTLFAFV
TH_0471|M.thermoresistible__bu RPLSKTGPRGTPVRITVLVGALVAIAASVFPVG--NLEEMVNIGTLFAFA
MAB_3599c|M.abscessus_ATCC_199 RELARTGSHGTPIRITLIVGFLVAVAASLTPIE--GLEEIVNVGTLFAFV
MLBr_01974|M.leprae_Br4923 TRLSSKNRFGVPIVAFGITAFLTLAFPLSLRFDSLNLTGLAVIARFIQFI
*: .. *.*: :.. . . . * :. :. :: *
Mb3281c|M.bovis_AF2122/97 LVSAGVVVLRRTRPD----LQRGFTAPWVPLLPIAAVCACLWLMLNLTAL
Rv3253c|M.tuberculosis_H37Rv LVSAGVVVLRRTRPD----LQRGFTAPWVPLLPIAAVCACLWLMLNLTAL
MMAR_1290|M.marinum_M LVSAGVIVLRKKRPD----LERGFRAPWVPVLPIASLCACLWLMLNLTAL
MUL_2597|M.ulcerans_Agy99 LVSAGVIVLRKKRPD----LERGFRAPWVPVLPIASLCACLWLMLNLTAL
MAV_4216|M.avium_104 LVSAGVMVLRRTRPD----LERGFKAPWVPVLPIASIGACVWLMLNLTAL
Mflv_4722|M.gilvum_PYR-GCK LVSAGVIVLRRTRPD----LKRGFRTPWVPVLPILSIIACVWLMLNLTGL
Mvan_1740|M.vanbaalenii_PYR-1 LVSAGVLVLRRTRPD----LKRGFRTPWVPLLPILAIAACVWLMLNLTGL
MSMEG_1838|M.smegmatis_MC2_155 LVSAGVLVLRRTRPD----LPRGFRTPLVPVLPVAAILACVWLMLNLTGL
TH_0471|M.thermoresistible__bu LVSAGVIVLRRTRPD----LKRGFRVPFVPWLPLAAIAACLWLMLNLTGL
MAB_3599c|M.abscessus_ATCC_199 LVSAGVIVLRRSRPD----LPRSFRTPGVPWLPIVAIVACGWLMLNLAVR
MLBr_01974|M.leprae_Br4923 IVPIALFALARSRTTNSVYVKRNVFTDKVLPITAVVVSAGLMVSFDYRSI
:*. .:..* :.*. : *.. . * :. : * : ::
Mb3281c|M.bovis_AF2122/97 TWIRFGIWLVAGTAIYVGYGRRHSAQGLRQARESATRRC----------
Rv3253c|M.tuberculosis_H37Rv TWIRFGIWLVAGTAIYVGYGRRHSAQGLRQARESATRRC----------
MMAR_1290|M.marinum_M TWIRFGVWLLVGTVIYVGYGRKHSMVGRRQLAEAAMDK-----------
MUL_2597|M.ulcerans_Agy99 TWIRFGVWLLVGTVIYVGYGRKHSMVGRRQLAEAAMDK-----------
MAV_4216|M.avium_104 TWVRFGIWLVVGTAIYAGYGYWHSVQGRRDAGEPDRSDALAADPNSPVP
Mflv_4722|M.gilvum_PYR-GCK TWIRFSVWMAIGVVLYFVYGRRHSLVARRQRERAGLHTDTP--------
Mvan_1740|M.vanbaalenii_PYR-1 TWIRFLIWMAIGVVVYFLYGRRHSLVGRRMG------------------
MSMEG_1838|M.smegmatis_MC2_155 TWIRFLIWMAIGVVIYFLYGRRRSVLATRDTTPTA--------------
TH_0471|M.thermoresistible__bu TWVRFLGWMALGLVLYFFYGRHHSMLGRGMTDISARRTG----------
MAB_3599c|M.abscessus_ATCC_199 TWLFALSWMVIGVIVYFAYSRKHSVLGLRVDVEQ---------------
MLBr_01974|M.leprae_Br4923 FLTRGGPNYFSISLIAITFIGIPAVAYLHYYRTSGIK------------
: : :