For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NMTQYAVTDPATAEVVATYPTATDAEVQAAIEAAHKTGRTWAKSTTVAERAALIRKVAELHEQRREQLAD VIVREMGKPRDQSLGEVDFCAAIYQYYADNAEKFLADEPIELLDGEGSAVIKRSPIGVLLGIMPWNYPYY QVARFAGPNLVVGNTILLKHAPQCPESAELLQLLFLEAGFPQGAYVDIRATNEQVANIIADPRVAAVSLT GSERAGAAVAEIAGRNLKKCVLELGGSDPFIVLSTDDLDATVQAAVDGRMENTGQACNAAKRFIVNESIY DDFLAKFTEKLLAAGEGIAPLSSEQAAVRLEDQVKSAVEAGAKLTAAGERKGAFFPPAVLTGVTEDNPVY RQELFGPVAVVYKAGSEEEAVQIANDTPFGLGSYVFTTDSEQAARVADKIEAGMVFVNAVGAEGAELPFG GVKRSGFGRELGRYGIDEFVNKKLIRVVD*
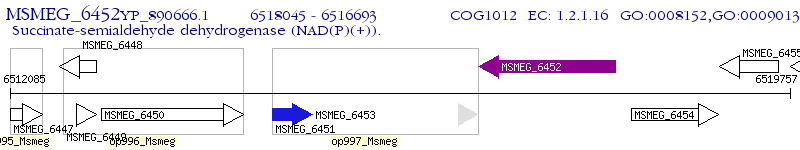
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6452 | - | - | 100% (450) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium smegmatis str. MC2 155] |
| M. smegmatis MC2 155 | MSMEG_6702 | - | e-126 | 54.12% (449) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium |
| M. smegmatis MC2 155 | MSMEG_2488 | - | 5e-70 | 36.30% (460) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0239c | gabD1 | 1e-105 | 46.02% (452) | succinic semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1858 | - | 1e-66 | 33.26% (454) | succinate-semialdehyde dehydrogenase (NAD(P)(+)) |
| M. tuberculosis H37Rv | Rv0234c | gabD1 | 1e-105 | 46.02% (452) | succinic semialdehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 1e-100 | 44.03% (452) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4322 | - | 0.0 | 76.06% (447) | aldehyde dehydrogenase family protein |
| M. marinum M | MMAR_0490 | gabD1 | 1e-103 | 44.93% (454) | succinate-semialdehyde dehydrogenase |
| M. avium 104 | MAV_4936 | gabD1 | 1e-105 | 45.58% (452) | succinic semialdehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_4137 | - | 1e-73 | 34.78% (460) | PUTATIVE putative [NADP+] succinate-semialdehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_1153 | gabD1 | 1e-104 | 45.15% (454) | succinic semialdehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1844 | - | 1e-123 | 53.33% (450) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6452|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4322|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_1844|M.vanbaalenii_PYR-1 -------------------------------------------------M
MMAR_0490|M.marinum_M --------------------------------------------------
MUL_1153|M.ulcerans_Agy99 --------------------------------------------------
MAV_4936|M.avium_104 --------------------------------------------------
Mb0239c|M.bovis_AF2122/97 MRSVTCSATLVLPVIEPTPADRRPRHLLLGSAGHVSGRLDTGRFVQTHPA
Rv0234c|M.tuberculosis_H37Rv MRSVTCSATLVLPVIEPTPADRRPRHLLLGSAGHVSGRLDTGRFVQTHPA
MLBr_02573|M.leprae_Br4923 --------------------------------------------------
Mflv_1858|M.gilvum_PYR-GCK -------------------------MDNSAVEALLASVPTGLWIGGEERE
TH_4137|M.thermoresistible__bu -----------------------------------------MYIDGEWQS
MSMEG_6452|M.smegmatis_MC2_155 -MNMTQYAVTDPATAEVVATYPTATDAEVQAAIEAAHKTGRTWAKSTTVA
MAB_4322|M.abscessus_ATCC_1997 ---MTEYAVTDPATGDVVATYPTATDAELDAALTAAAENSRTWPTDTTVA
Mvan_1844|M.vanbaalenii_PYR-1 TATQPPYRTLNPATGEVLATFDTATDDLIATAITAADDAYRAWR-EQPLE
MMAR_0490|M.marinum_M ----MPIATTNPATGETVKTFTAASNDEVDAAIARAYARFQDYRRNTTLA
MUL_1153|M.ulcerans_Agy99 ----MPIATTNPATGETVKTFTAASNDEVDAAIARAYARFQDYRRNTTFA
MAV_4936|M.avium_104 ----MPIATINPATGETVKTFTPASDAEVDAAIARAYERFLDYRHSTTFA
Mb0239c|M.bovis_AF2122/97 KDVSVPIATINPATGETVKTFTAATDDEVDAAIARAHRRFADYRQ-TSFA
Rv0234c|M.tuberculosis_H37Rv KDVSVPIATINPATGETVKTFTAATDDEVDAAIARAHRRFADYRQ-TSFA
MLBr_02573|M.leprae_Br4923 ----MSIATINPATGETVKTFTPTTQDEVEDAIARAYARFDNYRH-TTFT
Mflv_1858|M.gilvum_PYR-GCK GTS--TFDVLDPSDDTVLTSVADATADDARAALDAACAVQAEWAG-TAPR
TH_4137|M.thermoresistible__bu AASGKTFASINPANGAVLDSVPDGEREDARRAIDAAAAALPDWRS-RTAY
:*: .: : *: * : .
MSMEG_6452|M.smegmatis_MC2_155 ERAALIRKVAELHEQRREQLADVIVREMGKPRDQSLGEVDFCAAIYQYYA
MAB_4322|M.abscessus_ATCC_1997 DRAAQMRRVAQLHTEKREMLADIIVREMGKPRDEALGEIDFCAAIYEYYA
Mvan_1844|M.vanbaalenii_PYR-1 ARVDVVRHVGRLLRERATELAELAKLEMGKKLDEGTGEVEFSASIFEFYA
MMAR_0490|M.marinum_M QRAEWAHATADLIEAEADQTAALMTLEMGKTIASAKAEVLKSAKGFRYYA
MUL_1153|M.ulcerans_Agy99 QRAEWAHATADLIEAEADQTAALMTLEMGKTIASAKAEVLKSAKGFRYYA
MAV_4936|M.avium_104 QRAQWANATADLLEAEADEVAAMMTLEMGKTLKSAKAEALKCAKGFRYYA
Mb0239c|M.bovis_AF2122/97 QRARWANATADLLEAEADQAAAMMTLEMGKTLAAAKAEALKCAKGFRYYA
Rv0234c|M.tuberculosis_H37Rv QRARWANATADLLEAEADQAAAMMTLEMGKTLAAAKAEALKCAKGFRYYA
MLBr_02573|M.leprae_Br4923 QRAKWANATADLLEAEANQSAALMTLEMGKTLQSAKDEAMKCAKGFRYYA
Mflv_1858|M.gilvum_PYR-GCK KRGEILRSVFETITARADDIAALMTLEMGKVLAESKGEVTYGAEFFRWFA
TH_4137|M.thermoresistible__bu ERAEVLVEAHRIMLERRDDLAELMTREQGKPLRMARTEVGYAADFLLWFA
* . . * : * ** . * * ::*
MSMEG_6452|M.smegmatis_MC2_155 DNAEKFLADEPIEL-LDGEGSAVIKRSPIGVLLGIMPWNYPYYQVARFAG
MAB_4322|M.abscessus_ATCC_1997 DNAEKFLADERIEL-LAGEGTAVIKRFPIGVLLGIMPWNFPYYQVARFAC
Mvan_1844|M.vanbaalenii_PYR-1 DHAAALLADQRIPS-FSG-GTAVVQRRPVGVLLGVMPWNFPVYQVARFAA
MMAR_0490|M.marinum_M DNAAALLADEPADAGKVGASQAYTRYQPLGVVLAVMPWNFPLWQAVRFAA
MUL_1153|M.ulcerans_Agy99 DNAAALLADEPADAGKVGASQAYTRYQPLGVVLAVMPWNFPLWQAGRFAA
MAV_4936|M.avium_104 QNAEQLLADEPADAGKVGAARAYIRYQPLGVVLAVMPWNFPLWQAVRFAA
Mb0239c|M.bovis_AF2122/97 ENAEALLADEPADAAKVGASAAYGRYQPLGVILAVMPWNFPLWQAVRFAA
Rv0234c|M.tuberculosis_H37Rv ENAEALLADEPADAAKVGASAAYGRYQPLGVILAVMPWNFPLWQAVRFAA
MLBr_02573|M.leprae_Br4923 EKAETLLADEPAEAAAVGASKALARYQPLGVVLAVMPWNFPLWQTVRFAA
Mflv_1858|M.gilvum_PYR-GCK EEAVRIDG--RYTAAPAGTGRILVTKQAVGPCYAITPWNFPLAMGTRKMG
TH_4137|M.thermoresistible__bu EEAKRVYG--EVIPSARADQRFIVLAQPVGVCAAITPWNYPISMLTRKLA
:.* . . . .:* .: ***:* *
MSMEG_6452|M.smegmatis_MC2_155 PNLVVGNTILLKHAPQCPESAELLQLLFLEAGFPQGAYVDIRATNE--QV
MAB_4322|M.abscessus_ATCC_1997 PNLLVGNTIVLKHAPQCPESAEALALLFAEAGLPAGAYLNIRAANE--QV
Mvan_1844|M.vanbaalenii_PYR-1 PNLVLGNTILLKHAESVPQCALVIADVMRDAGVPEGVYTNLFASFD--QV
MMAR_0490|M.marinum_M PALMAGNVGLLKHASNVPQSALYLADVIARGGFPDGCFQTLLVSAS--AV
MUL_1153|M.ulcerans_Agy99 PALMAGNVGLLKHASNVPQSALYLADVIARAGFPDGCFQTLLVSAS--AV
MAV_4936|M.avium_104 PALMAGNVGILKHASNVPQSALYLADVITRGGFPEGCFQTLLVPSS--AV
Mb0239c|M.bovis_AF2122/97 PALMAGNVGLLKHASNVPQCALYLADVIARGGFPDGCFQTLLVSSG--AV
Rv0234c|M.tuberculosis_H37Rv PALMAGNVGLLKHASNVPQCALYLADVIARGGFPDGCFQTLLVSSG--AV
MLBr_02573|M.leprae_Br4923 PALMAGNVGLLKHASNVPQCALYLADVIARGGFPEDCFQTLLISAN--GV
Mflv_1858|M.gilvum_PYR-GCK PAFAAGCTMIVKPAQETPLTMLLLAKLMADAGLPKGVLSVLPTTSPGPVT
TH_4137|M.thermoresistible__bu PALAAGCTSVLKPAEQTPLCAVETFKVFADAGVPAGVVNLVTCSDPEPVA
* : * . ::* * . * :: .*.* . : . .
MSMEG_6452|M.smegmatis_MC2_155 ANIIADPRVAAVSLTGSERAGAAVAEIAGRNLKKCVLELGGSDPFIVLST
MAB_4322|M.abscessus_ATCC_1997 AGLIADPRVAGVSFTGSERAGAAVAEIAGRNLKKCVLELGGSDPFLVLSS
Mvan_1844|M.vanbaalenii_PYR-1 ETIIGDPRVQGVSVTGSERAGAAVAATAGRSLKKCLLELGGSDPFIVLD-
MMAR_0490|M.marinum_M EGILRDPRVAAATLTGSEPAGQSVGAIAGDEIKPTVLELGGSDPFIVMPS
MUL_1153|M.ulcerans_Agy99 EGILRDPRVAAATLTGSEPAGQSVGAIAGDEIKPTVLELGGSDPFIVMPS
MAV_4936|M.avium_104 ERILRDPRVAAATLTGSEPAGQSVAAIAGDEIKPTVLELGGSDPFIVMPS
Mb0239c|M.bovis_AF2122/97 EAILRDPRVAAATLTGSEPAGQSVGAIAGNEIKPTVLELGGSDPFIVMPS
Rv0234c|M.tuberculosis_H37Rv EAILRDPRVAAATLTGSEPAGQSVGAIAGNEIKPTVLELGGSDPFIVMPS
MLBr_02573|M.leprae_Br4923 EAILRDPRVAAATLTGSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPS
Mflv_1858|M.gilvum_PYR-GCK ETLIDDGRLRKLTFTGSTGVGKALVKQSADKLLRTSMELGGNAPFVVFDD
TH_4137|M.thermoresistible__bu DELLGDPRVRKISFTGSTEVGRMIASRAGATMKRVSMELGGHAPFLVFPD
:: * *: :.*** .* : :. : :**** **:*:
MSMEG_6452|M.smegmatis_MC2_155 DDLDATVQAAVDGRMENTGQACNAAKRFIVNESIYDDFLAKFTEKL--LA
MAB_4322|M.abscessus_ATCC_1997 DDLDATVEAAAAARLDNTGQACNAAKRFIVVDELYDAFVEKFTATL--LA
Mvan_1844|M.vanbaalenii_PYR-1 GDSKQLARTAWEFRVYNMGQACNSNKRMIVTDDLYDDFVAELVTLAGDLA
MMAR_0490|M.marinum_M ADLDKAVSTAVTGRVQNNGQSCIAAKRFIAHADIYDAFVDKFVEQMSALT
MUL_1153|M.ulcerans_Agy99 ADLDKAVSTAVTGRVQNNGQSCIAAKRFIAHADIYDAFVDKFVEQMSALT
MAV_4936|M.avium_104 ADLDEAVKTAVTARVQNNGQSCIAAKRFIVHTDIYDTFVDKFVEQMKALK
Mb0239c|M.bovis_AF2122/97 ADLDAAVSTAVTGRVQNNGQSCIAAKRFIVHADIYDDFVDKFVARMAALR
Rv0234c|M.tuberculosis_H37Rv ADLDAAVSTAVTGRVQNNGQSCIAAKRFIVHADIYDDFVDKFVARMAALR
MLBr_02573|M.leprae_Br4923 ADLDAAVATAVTGRVQNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLR
Mflv_1858|M.gilvum_PYR-GCK ADVDAAVDGAILAKMRNGGEACTAANRIHVANAVREEFTDKFVKRMSEFT
TH_4137|M.thermoresistible__bu ADPEHAAKGGAAVKFLNTGQACICPNRFIVHRSIAEPFIATLRARIEKLV
* . . . :. * *::* . :*: . : : * : :
MSMEG_6452|M.smegmatis_MC2_155 AGEG------IAPLSSEQAAVRLEDQVKSAVEAGAKLTAAGERKG-----
MAB_4322|M.abscessus_ATCC_1997 AADG------IAPLSSVRAAQTLEAQVAAAKAGGATVISAGERRG-----
Mvan_1844|M.vanbaalenii_PYR-1 PAE-------IAPMSSRKAAETLAAQVADAVDKGATLHVGGELAEGP---
MMAR_0490|M.marinum_M VGDPTDPQTQVGPLATEQSRDEIAQQVDDAAAAGAVIRCGGKPLAGP---
MUL_1153|M.ulcerans_Agy99 VGDPTDPQTQVGPLATEQSRDEIAQQVDDAAAAGAVIRCGGKPLAGP---
MAV_4936|M.avium_104 VGDPTDPATDVGPLATESGRDEIAKQVDDAVAAGATLRCGGKPLDGP---
Mb0239c|M.bovis_AF2122/97 VGDPTDPDTDVGPLATEQGRNEVAKQVEDAAAAGAVIRCGGKRLDRP---
Rv0234c|M.tuberculosis_H37Rv VGDPTDPDTDVGPLATEQGRNEVAKQVEDAAAAGAVIRCGGKRLDRP---
MLBr_02573|M.leprae_Br4923 VGDPTDPDTDVGPLATESGRAQVEQQVDAAAAAGAVIRCGGKRPDRP---
Mflv_1858|M.gilvum_PYR-GCK LGKGLDEKSTLGPLINAKQVATVTELVSDAVSRGATVAVGGVAPGGP---
TH_4137|M.thermoresistible__bu PGDGLAAGTTVGPLIDEVALKKMQRQVDDAVDKGAKLELGGSRLQGPAFD
.. :.*: : * * ** : .*
MSMEG_6452|M.smegmatis_MC2_155 --AFFPPAVLTGVTEDNPVYRQELFGPVAVVYKAGSEEEAVQIANDTPFG
MAB_4322|M.abscessus_ATCC_1997 --AFHPPAVITGLTADNPAYGQELFGPVASVYRASSEDQAVAIANDTPFG
Mvan_1844|M.vanbaalenii_PYR-1 -AAFYTPAVLTGVTPDMRAYSEELFGPVAVVYRVRDEEEAVALANASAYG
MMAR_0490|M.marinum_M -GWYYPPTVITDITKDMNLYTEEVFGPVASVYRAADIDEAIEIANATTFG
MUL_1153|M.ulcerans_Agy99 -GWYYPPTVITDITKDMNLYTEEVFGPVASVYRAADIDEAIEIANATTFG
MAV_4936|M.avium_104 -GWFYPPTVVTDITKDMALYTEEVFGPVASMYRAADIDEAIEIANATTFG
Mb0239c|M.bovis_AF2122/97 -GWFYPPTVITDISKDMALYTEEVFGPVASVFRAANIDEAVEIANATTFG
Rv0234c|M.tuberculosis_H37Rv -GWFYPPTVITDISKDMALYTEEVFGPVASVFRAANIDEAVEIANATTFG
MLBr_02573|M.leprae_Br4923 -GWFYPPTVITDITKDMALYTDEVFGPVASVYCATNIDDAIEIANATPFG
Mflv_1858|M.gilvum_PYR-GCK -GNFYPATVLTDVPADARILKEEVFGPVAPIAGFDTEEEGIAAANDTEYG
TH_4137|M.thermoresistible__bu NGHFYAPTILTGVTPDMLICSEETFGPIAPVMVFDDEDEAIAMANSTEYG
:...:::*.:. * :* ***:* : ::.: ** : :*
MSMEG_6452|M.smegmatis_MC2_155 LGSYVFTTDSEQAARVADKIEAGMVFVNAVGAEGAELPFGGVKRSGFGRE
MAB_4322|M.abscessus_ATCC_1997 LGSYVFTTDPEQAQRVANRIEAGMVFVNGVGADGAELPFGGIKRSGFGRE
Mvan_1844|M.vanbaalenii_PYR-1 LGASVFSTDTARAEKVAQRLEAGMVNVNTPAGEGPDIPFGGVKRSGFGRE
MMAR_0490|M.marinum_M LGSNAWTQDEAEQRRFINDIEAGQVFINGMTVSYPELPFGGIKRSGYGRE
MUL_1153|M.ulcerans_Agy99 LGSNAWTQDEAEQRRFINDIEAGQVFINGMTVSYPELPFGGIKRSGYGRE
MAV_4936|M.avium_104 LGSNAWTNDAAEQQRFIDDIEAGQVFINGMTVSYPELGFGGVKRSGYGRE
Mb0239c|M.bovis_AF2122/97 LGSNAWTRDETEQRRFIDDIVAGQVFINGMTVSYPELPFGGVKRSGYGRE
Rv0234c|M.tuberculosis_H37Rv LGSNAWTRDETEQRRFIDDIVAGQVFINGMTVSYPELPFGGVKRSGYGRE
MLBr_02573|M.leprae_Br4923 LGSNAWTQNESEQRHFIDNIVAGQVFINGMTVSYPELPFGGIKRSGYGRE
Mflv_1858|M.gilvum_PYR-GCK LAAYVYTQSLDRALRVAEGIESGMVGVNRGVISDPAAPFGGIKESGFGRE
TH_4137|M.thermoresistible__bu LAGYIYTRDISRAIRVGEALDFGMIGINDINPTSAAVPFGGIKQSGLGRE
*.. :: . . :. : : * : :* . ***:*.** ***
MSMEG_6452|M.smegmatis_MC2_155 LGRYGIDEFVNKKLIRVVD-------------
MAB_4322|M.abscessus_ATCC_1997 LGRFGMEEFVNKKLIRTL--------------
Mvan_1844|M.vanbaalenii_PYR-1 LGALGIDEFVNKQVYVVAD-------------
MMAR_0490|M.marinum_M LAGHGIREFCNIKTVWVG--------------
MUL_1153|M.ulcerans_Agy99 LAGHGIREFCNIKTVWVG--------------
MAV_4936|M.avium_104 LAGLGIRAFCNAKTVWIGSSKSGDAGGGSKVE
Mb0239c|M.bovis_AF2122/97 LSAHGIREFCNIKTVWIA--------------
Rv0234c|M.tuberculosis_H37Rv LSAHGIREFCNIKTVWIA--------------
MLBr_02573|M.leprae_Br4923 LSTHGIREFCNIKTVWIA--------------
Mflv_1858|M.gilvum_PYR-GCK GGTEGIEEYLDTKYIALTK-------------
TH_4137|M.thermoresistible__bu GARAGIEEYLDTKVLGIAL-------------
. *: : : :