For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDARILVVGAGAIGGVTAAHLARAGRDVTVLDANAEHVRLMNSPGLKFDELGETSHVTITAVDSPAALTG RFDFALVALKAPYLQAALGPLVERGLVDTYVSLGNGLVQNTIQQIVGTERFVVGITEWGATNLGPGHVAQ TTVAPFVIGEVDGTLTARLETLRGVLSAAAEVGVSTDVMNQVWTKLLLNSTFSGLAGVTGMLYGEIAADP LGRTMAYHVWTESYDVARAAGFEVGPLIGIQPDDLVVRSEADRERADAAIEVLMSKLGATKASMLQDLER GATTEVDVINGGVCTTAAKAGVASPFNARIVELVHECEQGLRSPGRETLEALAQISARQ*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
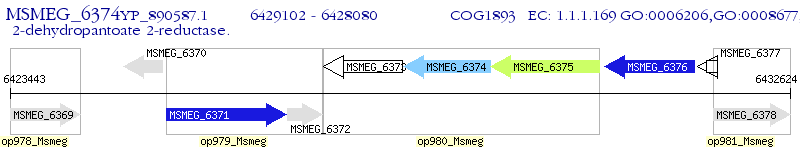
| M. smegmatis MC2 155 | MSMEG_6374 | panE | - | 100% (340) | 2-dehydropantoate 2-reductase |
| M. smegmatis MC2 155 | MSMEG_0153 | panE | 3e-17 | 26.52% (328) | 2-dehydropantoate 2-reductase |
| M. smegmatis MC2 155 | MSMEG_2919 | - | 5e-11 | 27.57% (341) | 2-dehydropantoate 2-reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0533 | - | 5e-15 | 27.49% (331) | ketopantoate reductase ApbA/PanE |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02430 | proC | 3e-06 | 25.42% (295) | pyrroline-5-carboxylate reductase |
| M. abscessus ATCC 19977 | MAB_2908c | - | 2e-11 | 26.18% (340) | 2-dehydropantoate 2-reductase |
| M. marinum M | MMAR_0863 | apbA | 3e-27 | 30.89% (327) | ketopantoate reductase, ApbA |
| M. avium 104 | MAV_4625 | panE | 7e-29 | 30.89% (327) | 2-dehydropantoate 2-reductase |
| M. thermoresistible (build 8) | TH_4612 | - | 4e-12 | 28.49% (358) | PUTATIVE putative 2-dehydropantoate 2-reductase |
| M. ulcerans Agy99 | MUL_0617 | apbA | 2e-23 | 30.58% (327) | ketopantoate reductase, ApbA |
| M. vanbaalenii PYR-1 | Mvan_0557 | - | 1e-15 | 27.79% (331) | ketopantoate reductase ApbA/PanE |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0863|M.marinum_M -----MKIAIIGCGAMGSIYAARLAGAGNDVAVIDRHQPNVDQITRTGLR
MUL_0617|M.ulcerans_Agy99 -----MKIAIIGCGAMGSIYAARLAGADNDVAVIDRHQPNVDQITRTGLR
MAV_4625|M.avium_104 -----MKIAVIGCGAMGSIYAAKLAAAGEQVLAVDRHRPSVERINRDGLR
Mflv_0533|M.gilvum_PYR-GCK -----MRVLILGAGGLGSVLGGYLANIGVDVTLVG-RPAHTEAITRYGLR
Mvan_0557|M.vanbaalenii_PYR-1 -----MRVLILGAGGLGSVLGGYLANIGVDVTLVG-RPAHAEAITRYGLR
MAB_2908c|M.abscessus_ATCC_199 -----MRIAVAGAGSIGCYVGGRLQAAGHQVVYIG-RPSLGRAIAEHGLS
MSMEG_6374|M.smegmatis_MC2_155 --MSDARILVVGAGAIGGVTAAHLARAGRDVTVLDANAEHVRLMNSPGLK
TH_4612|M.thermoresistible__bu --------VIVGSGAIGGTVGGLLARSGADVVLVA-RGAHAQALADRGLT
MLBr_02430|M.leprae_Br4923 MLSSMARIAIIGGGSIGEALLSGLLRAGRQVKDLVVAERMPDRARYLADT
: * *.:* . * . :* : .
MMAR_0863|M.marinum_M VSG-PGTDQLVS----LRAATTAPTEVMDLVVLAVKAADVAAGARQALC-
MUL_0617|M.ulcerans_Agy99 VSG-PGTDQLVS----LRAATTAPTEVMDLVVLAVKAADVAAGARQALC-
MAV_4625|M.avium_104 VTG-PGCDQVVP----LRASTGAVDETMDLVVLAVKAADVEAAARQALP-
Mflv_0533|M.gilvum_PYR-GCK ITG-RRGDLVITDNLTAVDQTAKATGHFDYLVLAVKGKDTAQALAEADG-
Mvan_0557|M.vanbaalenii_PYR-1 ITG-RRGDLVITDNLTAVDQTAKATGHFDYLVLAVKGKDTAQALAEADG-
MAB_2908c|M.abscessus_ATCC_199 LSDYLGWSAAIPASECAFSEDIDTVRSADIVLVTVKSAATAQMAESLAG-
MSMEG_6374|M.smegmatis_MC2_155 FDELGETSHVTIT---AVDSPAALTGRFDFALVALKAP-YLQAALGPLV-
TH_4612|M.thermoresistible__bu LRTPDGTFEIGVT--AAADPAEVRLTSRDVLVFATKTQHLEDALHAWVDR
MLBr_02430|M.leprae_Br4923 YSV-------------LVTSVTDAVENAMFVVVAVKPTDVESVMGDLVQ-
:.: *
MMAR_0863|M.marinum_M -------MLGADTPVLTIQNGLGSAETVADIVGEQRVAVGIASGFGAARL
MUL_0617|M.ulcerans_Agy99 -------MLGADTPVLTIQNGLGSAETVADIVGEQRVAVGIASGFGAARL
MAV_4625|M.avium_104 -------MLGPDTAVLTIQNGLGSAETVAGVVGAQRVAVGIASGFGASRV
Mflv_0533|M.gilvum_PYR-GCK -------LRDCVSSALSVQNTVEKDATLAGWLGPDRVIG-ASTIEGGTLL
Mvan_0557|M.vanbaalenii_PYR-1 -------LRDCVSSALSVQNTVEKDATLARWLGPDRVIG-ASTIEGGTLL
MAB_2908c|M.abscessus_ATCC_199 -------RLEPGAVVVSLQNGIRNPAQLSRYIKQHNVITGIVGFN---VA
MSMEG_6374|M.smegmatis_MC2_155 -------ERGLVDTYVSLGNGLVQN-TIQQIVGTERFVVGITEWG-ATNL
TH_4612|M.thermoresistible__bu PVHHGDTVVGTAGSTLPVLTALNGVVAEEMALRFFARVFGVCVWMPAAHV
MLBr_02430|M.leprae_Br4923 ------AAAVANDSAEQVLVTVAAGVTITYLESKLPAGTPVVRAMPNAAA
: :
MMAR_0863|M.marinum_M APG--HVHHNAMK------AMRFGAYSSLPHETVERIAHAWTEAGFDAAA
MUL_0617|M.ulcerans_Agy99 APGHVHVHHNAMK------AMRFGAYSSLPHETVERIAHAWTEVGFDAAA
MAV_4625|M.avium_104 APG--HVHHNAMR------AMRFGPYSSLPHARVESIARTWAHAGFDAAA
Mflv_0533|M.gilvum_PYR-GCK EPGLVRNHLTAEV------TAYFGELDGTSSARVDDLTAAFTNAGLPAKA
Mvan_0557|M.vanbaalenii_PYR-1 EPGLVRNHLTAEV------TAYFGELDGTSSARVDDLTAAFTNAGLPAKA
MAB_2908c|M.abscessus_ATCC_199 QVGAAHFHQGTQG------KLSLSTG-------AESLAGALTHGGLLTTV
MSMEG_6374|M.smegmatis_MC2_155 GPG--HVAQTTVA------PFVIGEVDGTLTARLETLRGVLS-AAAEVGV
TH_4612|M.thermoresistible__bu VPGEVIVRSWPVAGQFHISRWPADLADAEDDRLLSDIAATWTPAGLLVAR
MLBr_02430|M.leprae_Br4923 LVGAGVTVLAKGR-------FVTGQQFEDVLAMFDAVGGVLTVPESQMDA
* . . : . :
MMAR_0863|M.marinum_M VTDIAA--MQWEKLICNVAYSAP--CALTGMTVGQVLDDAELGP---VSR
MUL_0617|M.ulcerans_Agy99 VTDIAA--MQWEKLICNVAYSAP--CALTGMTVGQVLDDAEMGP---VSR
MAV_4625|M.avium_104 VTDIAA--MQWEKLICNVAYSAP--CALTGMTVGQVMDDAEMGP---VSR
Mflv_0533|M.gilvum_PYR-GCK VGNIEQ--VEWEKLAQIALAAAWSVTALGAIPDASVFEGMEVREGAMYFV
Mvan_0557|M.vanbaalenii_PYR-1 VGNIEQ--VEWEKLAQIALAAAWSVTALGAIPDASVFEGMEVREGAMYFV
MAB_2908c|M.abscessus_ATCC_199 RDDMAA--VQWGKLVVNLNNSVN---ALSGLPLKAELGDRDYRL---VLA
MSMEG_6374|M.smegmatis_MC2_155 STDVMN--QVWTKLLLNSTFSGL--AGVTGMLYGEIAADPLGRT---MAY
TH_4612|M.thermoresistible__bu PPDVAP--WKYRKLLANLANAVV---ALTGSGRGDPV----------VIA
MLBr_02430|M.leprae_Br4923 VTAVSGSGPAYFFLLVEALVDAGVAVGLTRQVATELAAQTMAGS-----A
: : * .:
MMAR_0863|M.marinum_M AAATEAWAVARASG----------------IAVAVTDPVAHARAFGAAMP
MUL_0617|M.ulcerans_Agy99 AAATEAWAVARASG----------------IAVAVTDPVAHARAFGAAMP
MAV_4625|M.avium_104 AAATEAWSVARASG----------------VAIAVADAVEHVRAFGAQMP
Mflv_0533|M.gilvum_PYR-GCK ALAKDLLGVYRAKGYEPMNFFAPLSRLKELDALPTADAIEFVIGQGRAMR
Mvan_0557|M.vanbaalenii_PYR-1 ALAKDLLGVYRAKGYEPMNFFAPLSRLKELDALPTADAIEFVIGQGRVMR
MAB_2908c|M.abscessus_ATCC_199 AAQREALGLLRRAG---QPVVSPLAAPLSLMPWVLTMPNPIFTRAARQMV
MSMEG_6374|M.smegmatis_MC2_155 HVWTESYDVARAAG---FEVGPLIGIQPDDLVVRSEADRERADAAIEVLM
TH_4612|M.thermoresistible__bu AAQREGEAVLRHAG-------------IEFVPAEVEREARRHGPRDRDVA
MLBr_02430|M.leprae_Br4923 AMLLERMDQDRHSA----------------EVAPLGAQVDVPAAQLRATI
: * .
MMAR_0863|M.marinum_M N----AKPSALLDHEARR-ASEIDVINGAVGRHGARVGVPTPVNTTLTAL
MUL_0617|M.ulcerans_Agy99 N----AKPSALLDHEARR-ASEIDVINGAVG-------VPTPVNTTLTAL
MAV_4625|M.avium_104 D----AKPSALLDHEARR-VSEIDVINGAVPRQGARVGVDAPVNATLTAV
Mflv_0533|M.gilvum_PYR-GCK EKGNSGHPSMYEDVLRRR-KTEVDFMLAPYLAAAQELGMDTPTLRVAYQI
Mvan_0557|M.vanbaalenii_PYR-1 EKGNLGHPSMYEDVLRRR-KTEVDFMLAPYLAAAQELGMDTPTLRVAYQI
MAB_2908c|M.abscessus_ATCC_199 AMDPQARSSMLDDITLGR-PTEIDYINGEVVELAKRLGAKAPVNAALVSL
MSMEG_6374|M.smegmatis_MC2_155 SKLGATKASMLQDLERGA-TTEVDVINGGVCTTAAKAGVASPFNARIVEL
TH_4612|M.thermoresistible__bu DAGDVVANSTWQSLARNSGSVETDYLNGEIVRIAHRHGMTAPVNAGLARL
MLBr_02430|M.leprae_Br4923 TSPGGTTAAALRELERGGLRMVVDAAVQAAKIRSEQLRITSE--------
: . * :
MMAR_0863|M.marinum_M VKSVERQWSAVNSD-------------------------
MUL_0617|M.ulcerans_Agy99 VKSVERQWSAVNSD-------------------------
MAV_4625|M.avium_104 VKSIERRWR------------------------------
Mflv_0533|M.gilvum_PYR-GCK IKTVDRFLA------------------------------
Mvan_0557|M.vanbaalenii_PYR-1 IKTVDRFLA------------------------------
MAB_2908c|M.abscessus_ATCC_199 IRDVTSSHEITRWPGPKLRREINHRAAASHMLS------
MSMEG_6374|M.smegmatis_MC2_155 VHECEQGLRSPGRETLEALAQISARQ-------------
TH_4612|M.thermoresistible__bu ARAAARDGHPPGRYSGGELAEALGLVGEALGADVMVEGR
MLBr_02430|M.leprae_Br4923 ---------------------------------------