For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPRDGNVWVIPAELSSAALVRNAAFEFAQLTLPAPSVGGATLRPVADRQDPLLHNMVDRIADLAARDDVL SRLLRDSIAEGLRLHIRDRYGEAPSPDGKSVQMLDQGQQRRLVEFLHDSLDERIDLRTAANIVGMNVRDF RPAFAQAFHTTLYQFVLDQRIKRAKALLASTALTMTEISFTVGFSSPSHFATTFKQRVGVTPSAYRAHL*
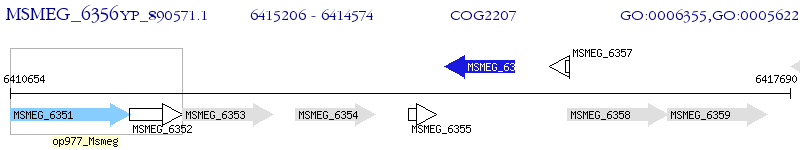
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6356 | - | - | 100% (210) | AraC family transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_0213 | - | 2e-10 | 32.11% (109) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_3998 | - | 7e-09 | 32.29% (96) | MmsAB operon regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1693 | - | 1e-08 | 31.93% (119) | AraC family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1313 | - | 2e-42 | 43.00% (207) | AraC family transcriptional regulator |
| M. marinum M | MMAR_1063 | - | 6e-07 | 29.07% (86) | transcriptional regulatory protein |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3120 | - | 6e-09 | 29.41% (153) | PUTATIVE transcriptional regulator, AraC family with |
| M. ulcerans Agy99 | MUL_0821 | - | 4e-07 | 29.07% (86) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3996 | - | 2e-13 | 23.53% (170) | helix-turn-helix domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6356|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1313|M.abscessus_ATCC_1997 ------MVFGGFGGVALGELQITFVSEPIVEPTEWYFDEPHHVVAVYQGG
Mflv_1693|M.gilvum_PYR-GCK MHTVAILAYDGMSGFESGLAAEIFGMTELSERFSAGLTRPWYTVKLCSES
TH_3120|M.thermoresistible__bu ---VAALVLDGLAIFEFGVICEVFGV----DRTDDGVPPVDFTVCGPRAG
MMAR_1063|M.marinum_M ------------MAQAARLIQHRSGVPIYGYRTDPDIPR--VSVLRPGPE
MUL_0821|M.ulcerans_Agy99 ------MDYLWRMAQAARLIQHRSGVPIYGYRTDPDIPR--VSVRRPGPE
Mvan_3996|M.vanbaalenii_PYR-1 ------MDATGVDRRVPARLVLATRDGQLPGADLRVYQFHSHQTQVPPLR
MSMEG_6356|M.smegmatis_MC2_155 ----------------MTPRDGNVWVIPAELS-----------SAALVRN
MAB_1313|M.abscessus_ATCC_1997 RILSKEIDFVDGLSRRYLPKVGDVLVVPMGSR-----------AAITTQG
Mflv_1693|M.gilvum_PYR-GCK AELRLLGGATVRTAYDLSDLAEADTVVIPSVRDIGDPVSSELVDAIKTAD
TH_3120|M.thermoresistible__bu QPVRTSFGATLTPDSGLDGLAGADLVAIPAIR--QDDYLPEALDAVRAAA
MMAR_1063|M.marinum_M DLGDRARHIHDFPALWYVPAAGLVYIAAAGQV----------LEPTQFGS
MUL_0821|M.ulcerans_Agy99 DLGDRARHIHDFPALWYVPAAGLVYIAAAGQV----------LEPTQFGS
Mvan_3996|M.vanbaalenii_PYR-1 DIAVIGWQSRAQIALRCGAIRSRAQMSPGDFS----ILRPGYESTWQWSR
: .
MSMEG_6356|M.smegmatis_MC2_155 AA-------FEFAQLTLPAPSVGGATLRPVADRQDPLLHNMVDR------
MAB_1313|M.abscessus_ATCC_1997 ER-------ASFCRFDIPTRLLDQRDLQPRVAYSDPLILQLASR------
Mflv_1693|M.gilvum_PYR-GCK ARGARLVSICSGAFALAAAGVLDGHKATTHWIYTDELRETYPGIDIDPNP
TH_3120|M.thermoresistible__bu DAGAIILTVCSGAFLAGAAGLLDGRPCTTHWGLTEELARRYPTARVDRNV
MMAR_1063|M.marinum_M LQGG------VAMFFDPAALGGDGGSPWPSWRTHPLLFPFLHGHSGGILR
MUL_0821|M.ulcerans_Agy99 LQAG------VAMFFDPAALGGDGGSPWPSWRTHPLLFPFLHGHSGGILR
Mvan_3996|M.vanbaalenii_PYR-1 GFQATVLYLNERRFADFASEVFDHHVDSVTIRESFQVQDSVIHHG-----
.: . :
MSMEG_6356|M.smegmatis_MC2_155 ----------IADLAARDDVLSRLLRDSIAEGLRLHIRDRYGEAPSPDGK
MAB_1313|M.abscessus_ATCC_1997 ----------MYSVADRTDVIARLLRESLADVVRLHLSDHY-AALRPRPE
Mflv_1693|M.gilvum_PYR-GCK LYVDNGRVLTSAGCAAGLDLCLHIVRTDHGVRVANDVARRLVISPHRAGG
TH_3120|M.thermoresistible__bu LYVDDGDLITSAGTAAGIDACLHLVRRELGSEVTNTIARHMVVPPQRDGG
MMAR_1063|M.marinum_M LEVPAARRSTWDGAIAAIEAELTARQQGYGQAVAAHLTLLLIELARLARD
MUL_0821|M.ulcerans_Agy99 LEVPAARRSTWDGAIAAIEAELTTRQQGYGQAVTAHLTLLLIELARLARD
Mvan_3996|M.vanbaalenii_PYR-1 -------LFLLATELRRDEFGAALYRDALVTQLCVHLLRHHAESRLRAHR
: : : :
MSMEG_6356|M.smegmatis_MC2_155 SVQMLDQ--------GQQRRLVEFLHDSLDERIDLRTAANIVGMNVRDFR
MAB_1313|M.abscessus_ATCC_1997 RRRLLDS--------AMQARVTEYIEDGLDADISLSALAGHAQMSTSEFR
Mflv_1693|M.gilvum_PYR-GCK QAQYIETPVPEPTEDGRIAAGMAWALEHLDETITLDELAARSAMSRRSYL
TH_3120|M.thermoresistible__bu QRQFIDQPIPVRHSEG-FAPHLDWILSNLDKTHTVASLARRANMSPRTFA
MMAR_1063|M.marinum_M VVGDLRR-----SGEPLLAEVFEVIDRRHAEPLSLRDVAAEVGMTPGHLT
MUL_0821|M.ulcerans_Agy99 VVGDLRR-----SGEPLLAEVFEVIDRRHAEPLSLRDVAAEVGMTPGHLT
Mvan_3996|M.vanbaalenii_PYR-1 HPGALNA--------AQAQQVTDYIEHNLDSDLCLSSLARVAGLSQYHFA
: : * :.
MSMEG_6356|M.smegmatis_MC2_155 PAFAQAFHTTLYQFVLDQRIKRAKALLASTALTMTEISFTVGFSSPSHFA
MAB_1313|M.abscessus_ATCC_1997 KAFTEVFGHTPYQFVLDRRMSRAKQLLATTDLSVTDISVAVGFSSPSHFA
Mflv_1693|M.gilvum_PYR-GCK RQFAKATGTTPIRWLIEQRIQASLALLESSDHSIDQIAARVGFESTATFR
TH_3120|M.thermoresistible__bu RRFVEETGRTPMQWVTDQRVLYARRLLEETDLDIHRIAARCGFGSTTLLR
MMAR_1063|M.marinum_M TIVRRRTGRTVQEWIIERRMAQARELLVATDLAVGEVARRVGISDPGYFS
MUL_0821|M.ulcerans_Agy99 TIVRRRTGRTVQEWIIERRMAQARELLVATDLAVGEVARRVGISDPGYFS
Mvan_3996|M.vanbaalenii_PYR-1 RLFKKRFGAPPHAYVQRRRIERARHLVLHSDMPLKEVVGVTGFCDQSHLT
. . . :: :*: : *: : : : *: . :
MSMEG_6356|M.smegmatis_MC2_155 TTFKQRVGVTPSAYRAHL------------------
MAB_1313|M.abscessus_ATCC_1997 TTFKTRIGVTPTAYRSGV------------------
Mflv_1693|M.gilvum_PYR-GCK HHFVRQMHTTPSDYR------SAFTGS---------
TH_3120|M.thermoresistible__bu HHFRRVIGVTPSDYRRRFACGSTTSGSTESGAEVTD
MMAR_1063|M.marinum_M RLFSRAHGTSPRQWRALPPRH---------------
MUL_0821|M.ulcerans_Agy99 RLFSRAHGTSPRQWRALPPRH---------------
Mvan_3996|M.vanbaalenii_PYR-1 RAFRRAFDITPAELRRSSD-----------------
* :* *